Here is another short excerpt from my recently published book, Neoplasms: Principles of Development and Diversity.
"Many more people are alive today as the result of precancer treatment than are alive due to the treatment of cancers.
The successful reduction in deaths from cervical cancer provides a good example of the effectiveness of precancer treatment. Cervical cancer is a type of squamous cell carcinoma that develops at the junction between the ectocervix (the squamous lined epithelium) and the endocervix (the glandular lined epithelium) in the os of the uterine cervix of women. Before the introduction of cervical precancer treatment, cervical carcinoma was one of the leading causes of cancer deaths in women. Today, in many countries that have not deployed precancer treatment, cervical cancer is the leading cause of cancer deaths in women (72, 73). The relatively low number of cervical cancer deaths in the United States is the result of a 70% reduction in age-adjusted mortality after the introduction of Pap smear screening (74–76). No effort aimed at treating invasive cancers has provided an equivalent reduction in the number of cancer deaths as this simple procedure for treating precancers.
Today, we know that almost all cervical cancer is due to infection by one of several carcinogenic strains of human papillomavirus. The strains of human papillomavirus that cause cervical cancer are transmitted during sexual intercourse by men infected with the virus. In the late 1940s (and really up until the early 1980s), the viral etiology of cervical cancer was unknown. We did know that morphologic changes in cervical epithelial characterized the early steps in cervical cancer development. By sampling and examining cervical specimens from women, it was possible to accurately determine whether precancerous changes were present. If precancerous changes were present, a gynecologist could remove a superficial portion of the affected epithelium, and this would, in the vast majority of cases, stop the cancer from ever developing.
Thanks largely to the persistence of Dr. Papanicolaou and his coworkers, a screening test was developed to detect cervical precancers. The Pap smear is obtained by scraping or brushing the junction between the endocervix and the ectocervix and
spreading the detached cells onto a glass slide. The cells are then stained with a histologic reagent (the Papanicolaou stain) that allows the cytologist to visualize subtle alterations in cell morphology. A typical Pap smear contains about 20,000 cells, and every cell must be inspected to rule out dysplasia and other pathologic abnormalities.
A large cytology laboratory can handle hundreds of thousands of Pap smears in a year. In the last half of the 20th century, the Pap smear cytologic evaluation led to at least a 70% drop in the number of deaths from cervical cancer in every country that fully deployed the test.
Morphologic and epidemiologic observations on Pap smears provided clues that led to the identification of several strains of human papillomavirus as the major causes of cervical cancer. [Half of the The 2008 Nobel Prize in Medicine went to Harald zur Hausen, for his work, showing the relationship between human papillomavirus and cervical cancer]. Recently, a vaccine against carcinogenic strains of HPV has been developed. Gardasil and Cervarix are two HPV vaccines that are currently available. If all goes well, most cases of cervical cancer will be prevented by a cancer vaccination. The Pap smear industry will shrink as fewer and fewer women test positive for cervical dysplasia.
The examination of Pap smears is just one of the many activities of a cytology laboratory. Cytologists routinely diagnose cells obtained from virtually any anatomic site and from any body fluid."
- Jules Berman
Devoted to the topic of data specification (including data organization, data description, data retrieval and data sharing) in the life sciences and in medicine.
Friday, November 28, 2008
Thursday, November 27, 2008
Thanks for all the data!
The United States is celebrating Thanksgiving today. Thanksgiving is my favorite holiday, because it allows everyone, regardless of their religion or personal philosophy, to join together as a nation in thanks for the many wonderful things this world provides.
This year is a particularly great Thanksgiving, because we have a newly elected President. Billions of people throughout the world hope that our new leadership will contribute to multinational efforts to reduce global warming, to use resources wisely and fairly, and to reduce violence and disease.
As someone who specializes in the field of biomedical informatics, I am personally grateful for the infrastructure that permits me to contribute to society, just by sitting at my computer and using the publicly available digital resources.
Here is the list of free resources that I am particularly grateful for (many of which accept charitable contributions):
Searchable information
PubMed
Wikipedia
Google
Google Earth
Languages and software
Perl
Ruby
Python
R
ImageMagick
OpenOffice
Datasets
SEER
Entrez
U.S. Census
Taxonomy.dat
OMIM
NASA images
Nomenclatures and ontologies
MESH
ICD
NCI Thesaurus
GO
Standards and protocols
HTML
XML
RDF
HTTP
FTP
JPEG
PNG
Organizations
GNU
OBO
CPAN
There are many many others, but these are all that I can think of at the moment. Please feel free to add your favorite free data resources, for which you are particularly thankful.
Happy Thanksgiving,
-Jules Berman
This year is a particularly great Thanksgiving, because we have a newly elected President. Billions of people throughout the world hope that our new leadership will contribute to multinational efforts to reduce global warming, to use resources wisely and fairly, and to reduce violence and disease.
As someone who specializes in the field of biomedical informatics, I am personally grateful for the infrastructure that permits me to contribute to society, just by sitting at my computer and using the publicly available digital resources.
Here is the list of free resources that I am particularly grateful for (many of which accept charitable contributions):
Searchable information
PubMed
Wikipedia
Google Earth
Languages and software
Perl
Ruby
Python
R
ImageMagick
OpenOffice
Datasets
SEER
Entrez
U.S. Census
Taxonomy.dat
OMIM
NASA images
Nomenclatures and ontologies
MESH
ICD
NCI Thesaurus
GO
Standards and protocols
HTML
XML
RDF
HTTP
FTP
JPEG
PNG
Organizations
GNU
OBO
CPAN
There are many many others, but these are all that I can think of at the moment. Please feel free to add your favorite free data resources, for which you are particularly thankful.
Happy Thanksgiving,
-Jules Berman
Sunday, November 23, 2008
Using SEER Public_use Data: 10
This is my tenth and final blog on the SEER Public-Use Data files.
I have prepared three web sites with much of the information that appeared in this series of 10 blog posts:
Obtaining the SEER data files and basic query subroutines, in Perl
A sample project
More examples
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics
More information on cancer is available in my recently published book, Neoplasms
© 2008 Jules Berman
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
I have prepared three web sites with much of the information that appeared in this series of 10 blog posts:
Obtaining the SEER data files and basic query subroutines, in Perl
A sample project
More examples
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics
More information on cancer is available in my recently published book, Neoplasms
© 2008 Jules Berman
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
Using SEER Public-Use Data: 9
I have been writing a series of blogs on SEER, the U.S. National Cancer Institute's Surveillance, Epidemiology and End Results program. SEER is an amazing resource for information on the cancers that occur in the U.S. One of the products of SEER is the Public Use dataset, which contains de-identified records on over 3.5 million cancers that have occurred between 1973 and 2005.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
Today, we'll look at the neoplasms that occur in the related anatomic sites: pleura, peritoneum, retro-peritoneum, and pelvis.
Here are the SEER listings. The left-hand column is the number of occurrences. Nneoplasms with fewer than 20 SEER cases were considered un-informative and were omitted from the lists. The second column is the average age of occurrence. The column on the right is the ICD-O term.
SEER captures malignant neoplasms. In the SEER data set, we saw that the following distribution of cases, by occurrences at anatomic site:
The pleura is the mesothelial-lined cavity of the chest, surrounding and covering the heart and the lungs. The peritoneum is the mesothelial-lined cavity of the abdomen, surrounding and overing all or part of the intestines and other organs of the abdomen (e.g., liver, spleen, pancreas).
The pleura accounts for many more occurrences of malignant tumors than does the peritoneum. Only a few different tumors account for the vast majority of malignant neoplasms arising in the pleura.
The peritoneum, with about half as many cancer occurrences as the pleura, has more types of tumors that occur in significant numbers (20 or more), including the tumors that arise from the surface of the ovaries (e.g. papillary serous cystadenocarcinoma).
The retroperitoneum, is the collection of tissues that lie between the peritoneal lining and the surface wall of the abdomen. The retroperitoneum is often referred to as the retroperitoneal space. This is not the best term, as it calls to mind a body cavity (space), perhaps lined by mesothelium, and this is not the case. The retroperitoneum is mostly fat, connective tissue, and organs. Fully retroperitoneal organs, such as the kidney and attached adrenals, can drop a little bit, along the potential space of its surronding fascia, but that's about the closest thing to a space that the retroperitoneum can offer. Organs of the abdomen that are slapped tightly against the posterior wall of the peritoneum are, technically, retroperitoneal (such as the ascending and descending colon, and the rectum). Organs or parts of organs that dangle in the abdomen (such as the transverse colon), are fully peritoneal.

For the purposes of collecting data on retroperitoneal neoplasms, the tumors that arise from identifiable organs (e.g., kidney, head of pancreas, adrenals, rectum) are assigned to those organs, in the SEER dataset, and NOT to the retroperitoneum. This leaves, for the most part, soft tissue tumors, muscle tumors and nerve tumors arising in the retroperitoneum. There are a great variety of these tumors, even when we restrict our list to those tumors that occur with a frequency of 20 or greater.
The pelvis is a commonly used anatomic term that creates much confusion. It is sometimes described as the bowl-like invagination in the lower abdomen, or it may be described as the structures that support the lower abdomen, or it may be described as the set of bones that create the framework of the bowl-like invagination.
It is very difficult to assign neoplasms to the pelvis, because tumors arising in this area can best be assigned to the bones in which they are found, or to the peritoneum, or to the retroperitoneum. The difficulty of assigning neoplasms to the pelvis becomes apparent when we see that of the approximately 3.5 million cases in the SEER dataset, there are only 470 cases assigned to the pelvis, and of these cases, most seem to arise, more specifically, from the peritoneum (papillary serous cystadenocarcinoma) or the uterine cervix (squamous cell carcinoma), or the intestines (adenocarcinoma).
There is a tautologic remark that pathologists use. "Common tumors occur commonly, and uncommon tumors occur uncommonly." This means that a pathologist should be cautious when assigning a diagnosis that rarely occurs at the location where the tumor has arisen.
To know which tumors commonly occur, at what sites, at what ages, in what ethnic populations, it is very useful to have a large collection of neoplasms from which to study, and to have a good understanding of the frequency of occurrence of the neoplasms that arise at the site. The SEER data set permits such determinations.
In a prior blog, I discussed some of the simple Perl routines used in the SEER-data parsing algorithms, and these are available from my web site.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics, myelodysplastic syndromes, IEN, pre-cancer, preneoplastic lesions, preneoplasia
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
The image of the retroperitoneum was taken from a reproduction of a Gray's Anatomy image, provided by Wikipedia. The copyright on Gray's Anatomy has expired, and the image is in the public domain.
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
In June, 2014, my book, entitled Rare Diseases and Orphan Drugs: Keys to Understanding and Treating the Common Diseases was published by Elsevier. The book builds the argument that our best chance of curing the common diseases will come from studying and curing the rare diseases.
I urge you to read more about my book. There's a generous preview of the book at the Google Books site. If you like the book, please request your librarian to purchase a copy of this book for your library or reading room.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
Today, we'll look at the neoplasms that occur in the related anatomic sites: pleura, peritoneum, retro-peritoneum, and pelvis.
Here are the SEER listings. The left-hand column is the number of occurrences. Nneoplasms with fewer than 20 SEER cases were considered un-informative and were omitted from the lists. The second column is the average age of occurrence. The column on the right is the ICD-O term.
SEER captures malignant neoplasms. In the SEER data set, we saw that the following distribution of cases, by occurrences at anatomic site:
PLEURA = 6,138 cases
PERITONEUM = 3,067 cases
RETROPERITONEUM = 3,640 cases
PELVIS = 470 cases
The pleura is the mesothelial-lined cavity of the chest, surrounding and covering the heart and the lungs. The peritoneum is the mesothelial-lined cavity of the abdomen, surrounding and overing all or part of the intestines and other organs of the abdomen (e.g., liver, spleen, pancreas).
The pleura accounts for many more occurrences of malignant tumors than does the peritoneum. Only a few different tumors account for the vast majority of malignant neoplasms arising in the pleura.
MALIGNANT NEOPLASMS OF THE PLEURA (MALE AND FEMALE)
TOTAL = 6,138 CASES
Number
of Age Neoplasm name
Cases
--------------------------------------------------------------
0024 068 sarcoma nos
0038 071 ml, large b-cell, diffuse
0110 070 neoplasm, malignant
0236 073 mesothelioma, biphasic type, malignant
0433 069 fibrous mesothelioma, malignant
1079 068 epithelioid mesothelioma, malignant
4027 069 mesothelioma, malignant
--------------------------------------------------------------
The peritoneum, with about half as many cancer occurrences as the pleura, has more types of tumors that occur in significant numbers (20 or more), including the tumors that arise from the surface of the ovaries (e.g. papillary serous cystadenocarcinoma).
NEOPLASMS OF THE PERITONEUM (MALE AND FEMALE)
TOTAL = 3067
Number
of Age Neoplasm name
Cases
--------------------------------------------------------------
022 062 liposarcoma nos
023 062 gastrointestinal stromal sarcoma
031 067 sarcoma nos
032 064 carcinoid tumor, malignant
038 085 endometrioid carcinoma
045 061 mucinous adenocarcinoma
046 066 fibrous histiocytoma, malignant
047 067 mullerian mixed tumor
053 066 neoplasm, malignant
073 072 carcinoma nos
073 063 ml, large b-cell, diffuse
097 068 papillary adenocarcinoma nos
127 063 leiomyosarcoma nos
134 062 epithelioid mesothelioma, malignant
180 065 serous cystadenocarcinoma nos
245 067 adenocarcinoma nos
359 066 serous surface papillary carcinoma
451 067 papillary serous cystadenocarcinoma
551 062 mesothelioma, malignant
--------------------------------------------------------------
The retroperitoneum, is the collection of tissues that lie between the peritoneal lining and the surface wall of the abdomen. The retroperitoneum is often referred to as the retroperitoneal space. This is not the best term, as it calls to mind a body cavity (space), perhaps lined by mesothelium, and this is not the case. The retroperitoneum is mostly fat, connective tissue, and organs. Fully retroperitoneal organs, such as the kidney and attached adrenals, can drop a little bit, along the potential space of its surronding fascia, but that's about the closest thing to a space that the retroperitoneum can offer. Organs of the abdomen that are slapped tightly against the posterior wall of the peritoneum are, technically, retroperitoneal (such as the ascending and descending colon, and the rectum). Organs or parts of organs that dangle in the abdomen (such as the transverse colon), are fully peritoneal.

For the purposes of collecting data on retroperitoneal neoplasms, the tumors that arise from identifiable organs (e.g., kidney, head of pancreas, adrenals, rectum) are assigned to those organs, in the SEER dataset, and NOT to the retroperitoneum. This leaves, for the most part, soft tissue tumors, muscle tumors and nerve tumors arising in the retroperitoneum. There are a great variety of these tumors, even when we restrict our list to those tumors that occur with a frequency of 20 or greater.
NEOPLASMS OF THE RETROPERITONEUM (MALE AND FEMALE)
TOTAL CASES = 3640
Number
of Age Neoplasm name
Cases
--------------------------------------------------------------
020 049 rhabdomyosarcoma nos
020 020 endodermal sinus tumor
022 021 teratoma, malignant nos
022 065 epithelioid leiomyosarcoma
023 010 embryonal rhabdomyosarcoma
024 069 mesothelioma, malignant
025 054 mesenchymoma, malignant
027 057 hemangiopericytoma, malignant
030 028 embryonal carcinoma nos
034 064 malignant lymphoma nos
035 048 neurofibrosarcoma
038 058 mixed type liposarcoma
040 066 malignant lymphoma, non hodgkin's type
043 045 neurilemmoma, malignant
045 044 seminoma nos
051 012 ganglioneuroblastoma
055 058 fibrosarcoma nos
060 066 pleomorphic liposarcoma
075 063 spindle cell sarcoma
111 063 dedifferentiated liposarcoma
127 067 neoplasm, malignant
140 062 myxoid liposarcoma
142 064 ml, large b-cell, diffuse
222 060 liposarcoma, well differentiated type
223 062 sarcoma nos
255 004 neuroblastoma nos
298 063 liposarcoma nos
311 063 fibrous histiocytoma, malignant
622 062 leiomyosarcoma nos
--------------------------------------------------------------
The pelvis is a commonly used anatomic term that creates much confusion. It is sometimes described as the bowl-like invagination in the lower abdomen, or it may be described as the structures that support the lower abdomen, or it may be described as the set of bones that create the framework of the bowl-like invagination.
It is very difficult to assign neoplasms to the pelvis, because tumors arising in this area can best be assigned to the bones in which they are found, or to the peritoneum, or to the retroperitoneum. The difficulty of assigning neoplasms to the pelvis becomes apparent when we see that of the approximately 3.5 million cases in the SEER dataset, there are only 470 cases assigned to the pelvis, and of these cases, most seem to arise, more specifically, from the peritoneum (papillary serous cystadenocarcinoma) or the uterine cervix (squamous cell carcinoma), or the intestines (adenocarcinoma).
NEOPLASMS OF THE PELVIS (MALE AND FEMALE)
TOTAL = 470
Number
of Age Neoplasm name
Cases
--------------------------------------------------------------
023 069 papillary serous cystadenocarcinoma
025 069 squamous cell carcinoma nos
050 075 carcinoma nos
061 066 adenocarcinoma nos
126 078 neoplasm, malignant
--------------------------------------------------------------
There is a tautologic remark that pathologists use. "Common tumors occur commonly, and uncommon tumors occur uncommonly." This means that a pathologist should be cautious when assigning a diagnosis that rarely occurs at the location where the tumor has arisen.
To know which tumors commonly occur, at what sites, at what ages, in what ethnic populations, it is very useful to have a large collection of neoplasms from which to study, and to have a good understanding of the frequency of occurrence of the neoplasms that arise at the site. The SEER data set permits such determinations.
In a prior blog, I discussed some of the simple Perl routines used in the SEER-data parsing algorithms, and these are available from my web site.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics, myelodysplastic syndromes, IEN, pre-cancer, preneoplastic lesions, preneoplasia
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
The image of the retroperitoneum was taken from a reproduction of a Gray's Anatomy image, provided by Wikipedia. The copyright on Gray's Anatomy has expired, and the image is in the public domain.
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
In June, 2014, my book, entitled Rare Diseases and Orphan Drugs: Keys to Understanding and Treating the Common Diseases was published by Elsevier. The book builds the argument that our best chance of curing the common diseases will come from studying and curing the rare diseases.
I urge you to read more about my book. There's a generous preview of the book at the Google Books site. If you like the book, please request your librarian to purchase a copy of this book for your library or reading room.
Saturday, November 22, 2008
Using SEER Public-Use Data: 8
I have been writing a series of blogs on SEER, the U.S. National Cancer Institute's Surveillance, Epidemiology and End Results program. SEER is an amazing resource for information on the cancers that occur in the U.S. One of the products of SEER is the Public Use dataset, which contains de-identified records on over 3.5 million cancers that have occurred between 1973 and 2005.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
Today, we'll look at the neoplasms that occur in appendix of the colon.
Here are the SEER listings. The left-hand column is the average age of occurrence of each neoplasm. The middle column is the number of cases in the SEER collection (neoplasms with fewer than 20 SEER cases were considered un-informative and were omitted from the list). The column on the right is the ICD-O term.
Though many different kinds of malignant neoplasms can occur in the appendix (and can be found in the SEER data), only carcinoids and adenocarcinomas occur frequently.
All of the carcinoid tumors cluster within a younger average age of occurrence than the adenocarcinomas.
This tells us a few things:
1. All of the carcinoids are biologically related to each other.
2. The carcinoids have a different developmental history than the adenocarcinomas.
3. When a pathologist sees a focus of adenocarcinoma in an appendiceal tumor, particularly in a young or middle-aged patient, he or she should carefully look for a focus of carcinoid, because the tumor might be a mixed adenocarcinoid tumor.
This is an example of how to use the SEER data to examine and test existing hypotheses and to develop new hypotheses. It took under a minute to generate the table, using a Perl script that parsed through 3.5 million SEER records.
In a prior blog, I discussed some of the simple Perl routines used in the SEER-data parsing algorithms, and these are available from my web site.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics, myelodysplastic syndromes, IEN, pre-cancer, preneoplastic lesions, preneoplasia
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
Today, we'll look at the neoplasms that occur in appendix of the colon.
Here are the SEER listings. The left-hand column is the average age of occurrence of each neoplasm. The middle column is the number of cases in the SEER collection (neoplasms with fewer than 20 SEER cases were considered un-informative and were omitted from the list). The column on the right is the ICD-O term.
Number
Age of Neoplasm name
Cases
--------------------------------------------------------------
039 022 carcinoid tumor, argentaffin, malignant
040 434 carcinoid tumor, malignant
050 217 adenocarcinoid tumor
054 224 mucocarcinoid tumor, malignant
055 038 composite carcinoid
058 022 carcinoma nos
058 138 signet ring cell carcinoma
059 186 mucin-producing adenocarcinoma
060 640 mucinous adenocarcinoma
061 175 mucinous cystadenocarcinoma nos
063 649 adenocarcinoma nos
065 033 adenocarcinoma in tubulovillous adenoma
067 063 adenocarcinoma in villous adenoma
--------------------------------------------------------------
Though many different kinds of malignant neoplasms can occur in the appendix (and can be found in the SEER data), only carcinoids and adenocarcinomas occur frequently.
All of the carcinoid tumors cluster within a younger average age of occurrence than the adenocarcinomas.
This tells us a few things:
1. All of the carcinoids are biologically related to each other.
2. The carcinoids have a different developmental history than the adenocarcinomas.
3. When a pathologist sees a focus of adenocarcinoma in an appendiceal tumor, particularly in a young or middle-aged patient, he or she should carefully look for a focus of carcinoid, because the tumor might be a mixed adenocarcinoid tumor.
This is an example of how to use the SEER data to examine and test existing hypotheses and to develop new hypotheses. It took under a minute to generate the table, using a Perl script that parsed through 3.5 million SEER records.
In a prior blog, I discussed some of the simple Perl routines used in the SEER-data parsing algorithms, and these are available from my web site.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics, myelodysplastic syndromes, IEN, pre-cancer, preneoplastic lesions, preneoplasia
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
Thursday, November 20, 2008
Using SEER Public-Use Data: 7
I have been writing a series of blogs on SEER, the U.S. National Cancer Institute's Surveillance, Epidemiology and End Results program. SEER is an amazing resource for information on the cancers that occur in the U.S. One of the products of SEER is the Public Use dataset, which contains de-identified records on over 3.5 million cancers that have occurred between 1973 and 2005.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
Yesterday, we used the SEER data to show that cervical in situ carcinomas, precancers that precede the development of invasive cancers of the cervix, occur in populations with an average age younger than the average age of occurrence of the advanced lesions (just as we expected).
Today, we'll look at the neoplasms that occur in the blood and bone marrow. Do the precancers of blood occur at a younger age than the cancers that develop from those precancers (as we might expect)?
Here are the SEER listings for neoplasms of the blood and bone marrow. The left-hand column is the average age of occurrence of each neoplasm. The middle column is the number of cases in the SEER collection (neoplasms with fewer than 20 SEER cases were considered un-informative and were omitted from the list). The column on the right is the ICD-O term.
The precancer lesions of the bood cells are the myelodysplasias (previously called preleukemias). They include the refractory anemias and chronic myelomyocytic leukemia (not to be confused with chronic myeloid leukemia). These lesions, sometimes progress to acute myelogenous leukemia.
Here are the average ages of development of the myelodysplasias:
All of the myelodysplasias cluster at the upper end of ages for blood neoplasms (70+ years old). This is far older than the average age of occurrence of the acute leukemias (into which the myelodysplasias develop).
How can a precursor lesions occur in a population that is older than the population in which the developed cancer occurs?
The answer is simple. Most acute leukemias do not develop from the myelodysplasias. When we look at column two, we see at a glance that the acute leukemias are much more numerous than the myelodysplasias.
The pathway of myelodysplasia to acute leukemia is the exception, not the rule, and we would need to find some other precursor lesion to accunt for the bulk of acute myeloid leukemias.
This is an example of how to use the SEER data to examine and test existing hypotheses and to develop new hypotheses. It took under a minute to generate the table, using a Perl script that parsed through 3.5 million SEER records.
In a prior blog, I discussed some of the simple Perl routines used in the SEER-data parsing algorithms, and these are available from my web site.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics, myelodysplastic syndromes, IEN, pre-cancer, preneoplastic lesions, preneoplasia
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
Yesterday, we used the SEER data to show that cervical in situ carcinomas, precancers that precede the development of invasive cancers of the cervix, occur in populations with an average age younger than the average age of occurrence of the advanced lesions (just as we expected).
Today, we'll look at the neoplasms that occur in the blood and bone marrow. Do the precancers of blood occur at a younger age than the cancers that develop from those precancers (as we might expect)?
Here are the SEER listings for neoplasms of the blood and bone marrow. The left-hand column is the average age of occurrence of each neoplasm. The middle column is the number of cases in the SEER collection (neoplasms with fewer than 20 SEER cases were considered un-informative and were omitted from the list). The column on the right is the ICD-O term.
Number
Age of Neoplasm name
Cases
--------------------------------------------------------------
015 0000057 langerhans cell histiocytosis, disseminated
021 0000827 precursor b-cell lymphoblastic leukemia
023 0009220 precursor cell lymphoblastic leukemia, nos
024 0000117 precursor t-cell lymphoblastic leukemia
042 0000117 burkitt cell leukemia
043 0000024 burkitt lymphoma, nos
045 0000233 burkitt's tumor
047 0001140 acute promyelocytic leukemia
048 0000257 megakaryocytic leukemia
048 0000057 ac. myelomonocytic leuk. w abn. mar. eosinophils
050 0000034 acute biphenotypic leukemia
050 0000085 acute myeloid leukemia, t(8;21)(q22;q22)
052 0000248 chronic myelogenous leukemia, bcr/abl positive
053 0000053 hypereosinophilic syndrome
054 0000121 malignant mastocytosis
055 0000032 megakaryocytic myelosis
055 0000031 mature t-cell lymphoma, nos
058 0002146 hairy cell leukemia
058 0001770 acute monocytic leukemia
059 0000030 hodgkin's disease nos
059 0000447 acute myeloid leukemia without maturation
059 0000022 acute myeloid leukemia, 11q23 abnormalities
060 0000115 myeloid sarcoma
060 0000614 acute myeloid leukemia with maturation
060 0000113 adult t-cell leukemia/lymphoma (htlv-1 pos.)
061 0018129 acute myeloid leukemia
061 0010279 chronic myeloid leukemia
061 0002006 acute myelomonocytic leukemia
061 0000324 acute myeloid leukemia, minimal differentiation
061 0000027 malignant lymphoma, mixed lymphocytic-histiocytic, nodular
062 0000086 therapy-related myelodysplastic syndrome, nos
063 0000032 hemangiosarcoma
063 0000119 plasma cell tumor, malignant
063 0000054 ml, large b-cell, diffuse, immunoblastic, nos
064 0001275 polycythemia vera
064 0000428 ml, large b-cell, diffuse
064 0000087 acute panmyelosis with myelofibrosis
065 0003731 acute leukemia nos
065 0000200 plasma cell leukemia
065 0000998 essential thrombocythemia
065 0000482 plasmacytoma, extramedullary
066 0000815 erythroleukemia
066 0000036 malignant lymphoma, nodular nos
066 0000036 ml, mixed sm. and lg. cell, diffuse
066 0000042 prolymphocytic leukemia, t-cell type
066 0000162 splenic marginal zone b-cell lymphoma
066 0000026 malignant lymphoma, follicular center cell, cleaved, follicular
067 0001014 lymphoid leukemia nos
067 0000225 malignant lymphoma, non hodgkin's type
067 0000327 acute myeloid leuk. with multilineage dysplasia
068 0000148 ml, lymphoplasmacytic
068 0000132 marginal zone b-cell lymphoma, nos
068 0000032 prolymphocytic leukemia, b-cell type
069 0036377 multiple myeloma
069 0001870 myeloid leukemia nos
069 0000094 mantle cell lymphoma
069 0000314 malignant lymphoma nos
069 0000362 myelosclerosis with myeloid metaplasia
069 0000586 chronic myeloproliferative disease, nos
070 0030307 chronic lymphoid leukemia
070 0000301 prolymphocytic leukemia, nos
071 0000276 ml, small b lymphocytic, nos
071 0001582 waldenstrom macroglobulinemia
071 0000263 refractory cytopenia with multilineage dysplasia
071 0000080 refract. anemia with excess blasts in transformation
072 0002580 leukemia nos
072 0000763 refractory anemia with excess blasts
073 0000820 refractory anemia
074 0002422 myelodysplastic syndrome, nos
074 0000655 refractory anemia with sideroblasts
074 0001798 chronic myelomonocytic leukemia, nos
074 0000089 myelodysplastic syndr. with 5q deletion syndrome
----------------------------------------------------------------
The precancer lesions of the bood cells are the myelodysplasias (previously called preleukemias). They include the refractory anemias and chronic myelomyocytic leukemia (not to be confused with chronic myeloid leukemia). These lesions, sometimes progress to acute myelogenous leukemia.
Here are the average ages of development of the myelodysplasias:
Number
Age of Neoplasm name
Cases
--------------------------------------------------------------
071 0000263 refractory cytopenia with multilineage dysplasia
071 0000080 refract. anemia with excess blasts in transformation
072 0000763 refractory anemia with excess blasts
073 0000820 refractory anemia
074 0002422 myelodysplastic syndrome, nos
074 0000655 refractory anemia with sideroblasts
074 0001798 chronic myelomonocytic leukemia, nos
074 0000089 myelodysplastic syndr. with 5q deletion syndrome
--------------------------------------------------------------
All of the myelodysplasias cluster at the upper end of ages for blood neoplasms (70+ years old). This is far older than the average age of occurrence of the acute leukemias (into which the myelodysplasias develop).
Number
Age of Neoplasm name
Cases
--------------------------------------------------------------
050 0000085 acute myeloid leukemia, t(8;21)(q22;q22)
058 0001770 acute monocytic leukemia
059 0000447 acute myeloid leukemia without maturation
059 0000022 acute myeloid leukemia, 11q23 abnormalities
060 0000614 acute myeloid leukemia with maturation
061 0018129 acute myeloid leukemia
061 0002006 acute myelomonocytic leukemia
061 0000324 acute myeloid leukemia, minimal differentiation
065 0003731 acute leukemia nos
067 0000327 acute myeloid leuk. with multilineage dysplasia
--------------------------------------------------------------
How can a precursor lesions occur in a population that is older than the population in which the developed cancer occurs?
The answer is simple. Most acute leukemias do not develop from the myelodysplasias. When we look at column two, we see at a glance that the acute leukemias are much more numerous than the myelodysplasias.
The pathway of myelodysplasia to acute leukemia is the exception, not the rule, and we would need to find some other precursor lesion to accunt for the bulk of acute myeloid leukemias.
This is an example of how to use the SEER data to examine and test existing hypotheses and to develop new hypotheses. It took under a minute to generate the table, using a Perl script that parsed through 3.5 million SEER records.
In a prior blog, I discussed some of the simple Perl routines used in the SEER-data parsing algorithms, and these are available from my web site.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics, myelodysplastic syndromes, IEN, pre-cancer, preneoplastic lesions, preneoplasia
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
Wednesday, November 19, 2008
Using SEER Public-Use Data: 6
I have been writing a series of blogs on SEER, the U.S. National Cancer Institute's Surveillance, Epidemiology and End Results program. SEER is an amazing resource for information on the cancers that occur in the U.S. One of the products of SEER is the Public Use dataset, which contains de-identified records on over 3.5 million cancers that have occurred between 1973 and 2005.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
Here is an example. Precancers are the identifiable, easily-treated, lesions from which advanced cancers develop. When you eradicate the precancer, the cancer never develops.
In theory, if precancers precede cancers, the average age at diagnosis of precancers should be smaller than the average at diagnosis of the cancers that develop from precancers. This biologic tautology has been hard to verify, because there is not all precancers will develop in people of the same age. The same is true for cancers. And it's hard to come up with a large enough population that will separate the (overlapping) precancer and cancer populations.
However, the SEER population provides the numbers we need. When we extract all of the SEER neoplasms that arise in the uterine cervix, we find the following average ages for the resulting set of tumors:
The average age of all of the in situ lesions (i.e., non-invasive precancers) is smaller than the average age of the observed invasive cancers arising from the cervix! I have never seen this observation demonstrated from any other data set.
It took about one minute to generate the table, using a Perl script that parsed through 3.5 million SEER records.
In yesterday's blog, I discussed some of the simple Perl routines used in the SEER-data parsing algorithms.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
Here is an example. Precancers are the identifiable, easily-treated, lesions from which advanced cancers develop. When you eradicate the precancer, the cancer never develops.
In theory, if precancers precede cancers, the average age at diagnosis of precancers should be smaller than the average at diagnosis of the cancers that develop from precancers. This biologic tautology has been hard to verify, because there is not all precancers will develop in people of the same age. The same is true for cancers. And it's hard to come up with a large enough population that will separate the (overlapping) precancer and cancer populations.
However, the SEER population provides the numbers we need. When we extract all of the SEER neoplasms that arise in the uterine cervix, we find the following average ages for the resulting set of tumors:
Number
Age of Neoplasm name
Cases
--------------------------------------------------------------
034 0049176 carcinoma in situ nos
034 0051906 squamous cell carcinoma in situ nos
035 0000359 sq cell carcinoma lg cell non-ker in situ
037 0000313 sq cell carcinoma keratinizing nos in situ
039 0001348 adenocarcinoma in situ
039 0018551 squamous intraepithelial neoplasia, grade iii
039 0000058 squamous cell carcinoma in situ with questionable stromal invasion
041 0003213 squamous cell carcinoma, microinvasive
043 0000113 adenocarcinoma, endocervical type
048 0001320 adenosquamous carcinoma
048 0000049 neuroendocrine carcinoma
048 0000093 large cell carcinoma nos
049 0003118 squamous cell carcinoma, large cell, nonkeratinizing type
050 0002524 carcinoma nos
050 0000259 endometrioid carcinoma
050 0000021 mucinous adenocarcinoma, endocervical type
051 0004121 adenocarcinoma nos
051 0000250 small cell carcinoma nos
051 0002727 squamous cell carcinoma, keratinizing type nos
051 0000104 squamous cell carcinoma, small cell, nonkeratinizing type
052 0000233 mucinous adenocarcinoma
052 0018774 squamous cell carcinoma nos
054 0000218 clear cell adenocarcinoma nos
055 0000088 papillary squamous cell carcinoma
056 0000023 mesodermal mixed tumor
056 0000141 mucin-producing adenocarcinoma
058 0000034 verrucous carcinoma nos
060 0000289 neoplasm, malignant
060 0000037 papillary serous cystadenocarcinoma
062 0000025 sarcoma nos
062 0000055 mullerian mixed tumor
062 0000044 carcinoma, anaplastic type nos
062 0000076 carcinoma, undifferentiated type nos
062 0000033 adenocarcinoma with squamous metaplasia
063 0000043 carcinosarcoma nos
----------------------------------------------------------------
The average age of all of the in situ lesions (i.e., non-invasive precancers) is smaller than the average age of the observed invasive cancers arising from the cervix! I have never seen this observation demonstrated from any other data set.
It took about one minute to generate the table, using a Perl script that parsed through 3.5 million SEER records.
In yesterday's blog, I discussed some of the simple Perl routines used in the SEER-data parsing algorithms.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
Tuesday, November 18, 2008
Using SEER Public Use Data: 5
SEER is the U.S. National Cancer Institute's Surveillance, Epidemiology and End Results program. It is an amazing resource for information about the cancers that occur in the U.S. One of the products of SEER is the Public Use dataset, which contains de-identified records on over 3.5 million cancers that have occurred between 1973 and 2005.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
The SEER public-use data files are available as a DVD or they can be downloaded from the web.
Information for obtaining these files is available at:
http://seer.cancer.gov/data/
To get these files, you need to fax a signed agreement to the National Library of Medicine and wait for a response (it just took a few hours when I tried a week or two ago).
For this exercise, I downloaded the following file:
SEER_1973_2005_CD2.d08042008.zip (199,212,317 bytes)
Expanded, it produced several directories of files. The raw data records are contained in the following files.
The SEER files comprise over 920 megabytes of text.
Each record looks something like this (note that the following is a fake and truncated record, because I didn't want to list an actual record from SEER on my web page. JB):
An actual record might go on for 258 characters.
Suppose we wanted to parse through every record of every file in the SEER data set.
A few lines of Perl will suffice:
The output of the script is the number "3553255" (the total number of parsed records). It will appear on your computer monitor, after about 11 seconds. That's all the time it takes (on a 2.8 GHz CPU) to parse 920 megabytes of text.
Each SEER record is a cancer case, described by a series of 258 (mostly) numbers, in byte-assigned positions, described by a data dictionary document (available at the SEER web site). Here are the first 14 items in the dictionary.
These first 14 items are the only items I have used for any of my SEER projects.
When you know the byte locations for the data dictionary entries, you can easily write a short script (I like to use Perl, Ruby, or Python) that can extract and compile data any way you wish.
Here's a short Perl script that parses through the first two records of each SEER public use file, extracting the age at diagnosis and the year of diagnosis from the first two records of each SEER file, and printing out the result to the screen.
The output looks like:
The key line from the Perl script is:
This pulls the string consisting of bytes 25,26, and 27 from the record. The data dictionary tells us that bytes 25-27 comprise the record's age at diagnosis.
Two of the most important data items in the record will require a little extra work, if, once extracted, you want to understand their meaning.
These are the morphology codes and the anatomic site codes.
The morphology code occupies bytes 53-57 (for ICDO-3) and bytes 48-52 (for ICDO-2), for each record.
Examples of morphology codes are:
You can download a copy of the ICDO-3 codes from SEER
If you start with a pdf file of the ICDO codes, you will need to cut and paste the pdf file into a plain ascii text file, free of formatting characters, to use it in your scripts. I put the list of codes and equivalent terms into a text file that I named "ICDO-3".
I often use the following short subroutine to put the codes, and their term equivalents, into a hash. When I need to convert a code into its term, I just call the hash value.
This snippet of code will mean something to you when you look at the data format in SEER's ICDO-3 file.
The same can be done with an ICDO-2 file, and with the anatomic site codes (the primary site item, bytes 43-46).
Anatomic site codes and terms are available at:
http://www.ncri.ie/data.cgi/html/icdo2sites.shtml
An example of a site code and its term equivalent is:
Everything we want to do with the SEER files involves parsing through the files, line (record) by line; and then pulling out the data we're interested in.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
In June, 2014, my book, entitled Rare Diseases and Orphan Drugs: Keys to Understanding and Treating the Common Diseases was published by Elsevier. The book builds the argument that our best chance of curing the common diseases will come from studying and curing the rare diseases.
I urge you to read more about my book. There's a generous preview of the book at the Google Books site. If you like the book, please request your librarian to purchase a copy of this book for your library or reading room.
tags: common disease, orphan disease, orphan drugs, rare disease, subsets of disease, disease genetics, genetics of complex disease, genetics of common diseases, neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
The SEER public-use data files are available as a DVD or they can be downloaded from the web.
Information for obtaining these files is available at:
http://seer.cancer.gov/data/
To get these files, you need to fax a signed agreement to the National Library of Medicine and wait for a response (it just took a few hours when I tried a week or two ago).
For this exercise, I downloaded the following file:
SEER_1973_2005_CD2.d08042008.zip (199,212,317 bytes)
Expanded, it produced several directories of files. The raw data records are contained in the following files.
08/04/2008 11:09 AM 143,352,097 BREAST.TXT
08/04/2008 11:09 AM 109,510,121 COLRECT.TXT
08/04/2008 11:09 AM 67,313,323 DIGOTHR.TXT
08/04/2008 11:09 AM 93,497,187 FEMGEN.TXT
08/04/2008 11:09 AM 70,809,823 LYMYLEUK.TXT
08/04/2008 11:09 AM 119,312,494 MALEGEN.TXT
08/04/2008 11:09 AM 127,835,148 OTHER.TXT
08/04/2008 11:09 AM 129,526,418 RESPIR.TXT
08/04/2008 11:09 AM 59,133,585 URINARY.TXT
9 File(s) 920,290,196 bytes
The SEER files comprise over 920 megabytes of text.
Each record looks something like this (note that the following is a fake and truncated record, because I didn't want to list an actual record from SEER on my web page. JB):
246000990000001521205001078191409902051986C64908130381303211
An actual record might go on for 258 characters.
Suppose we wanted to parse through every record of every file in the SEER data set.
A few lines of Perl will suffice:
#/usr/local/bin/perl
opendir(SEERDIR, "c\:\\seer") || die ("Unable to open directory");
@files = readdir(SEERDIR);
$totalcount;
closedir(SEERDIR);
chdir("c\:\\seer");
foreach $datafile (@files)
{
open (TEXT, $datafile);
$line = " ";
while ($line ne "")
{
$line = <TEXT>;
$totalcount++;
}
close TEXT;
}
print "\n$totalcount";
exit
The output of the script is the number "3553255" (the total number of parsed records). It will appear on your computer monitor, after about 11 seconds. That's all the time it takes (on a 2.8 GHz CPU) to parse 920 megabytes of text.
Each SEER record is a cancer case, described by a series of 258 (mostly) numbers, in byte-assigned positions, described by a data dictionary document (available at the SEER web site). Here are the first 14 items in the dictionary.
List of the data dictionary items accounting for the first 46 bytes of a SEER record
Patient ID number 01-08
Registry ID 09-18
Marital Status at DX 19-19
Race/Ethnicity 20-21
Spanish/Hispanic Origin 22-22
NHIA Derived Hispanic Origin 23-23
Sex 24-24
Age at diagnosis 25-27
Year of Birth 28-31
Birth Place 32-34
Sequence Number--Central 35-36
Month of diagnosis 37-38
Year of diagnosis 39-42
Primary Site 43-46
These first 14 items are the only items I have used for any of my SEER projects.
When you know the byte locations for the data dictionary entries, you can easily write a short script (I like to use Perl, Ruby, or Python) that can extract and compile data any way you wish.
Here's a short Perl script that parses through the first two records of each SEER public use file, extracting the age at diagnosis and the year of diagnosis from the first two records of each SEER file, and printing out the result to the screen.
#/usr/local/bin/perl
opendir(SEERDIR, "c\:\\seer") || die ("Unable to open directory");
@files = readdir(SEERDIR);
closedir(SEERDIR);
chdir("c\:\\seer");
foreach $datafile (@files)
{
next if ($datafile !~ /\.txt/i);
open (TEXT, $datafile);
for($i=0;$i<2;$i++)
{
$line = <TEXT>;
$age_at_dx = substr($line,24,3);
$year_of_dx = substr($line,38,4);
print "$datafile Age $age_at_dx Year $year_of_dx\n";
}
close TEXT;
}
exit
The output looks like:
BREAST.TXT Age 057 Year 1988
BREAST.TXT Age 094 Year 1979
COLRECT.TXT Age 059 Year 1973
COLRECT.TXT Age 069 Year 1989
DIGOTHR.TXT Age 062 Year 1981
DIGOTHR.TXT Age 062 Year 1978
FEMGEN.TXT Age 080 Year 2000
FEMGEN.TXT Age 067 Year 1983
LYMYLEUK.TXT Age 071 Year 1990
LYMYLEUK.TXT Age 078 Year 1990
MALEGEN.TXT Age 088 Year 1984
MALEGEN.TXT Age 082 Year 1979
OTHER.TXT Age 064 Year 1979
OTHER.TXT Age 067 Year 1983
RESPIR.TXT Age 074 Year 1977
RESPIR.TXT Age 091 Year 2004
URINARY.TXT Age 071 Year 1986
URINARY.TXT Age 074 Year 1973
The key line from the Perl script is:
$age_at_dx = substr($line,24,3);
This pulls the string consisting of bytes 25,26, and 27 from the record. The data dictionary tells us that bytes 25-27 comprise the record's age at diagnosis.
Two of the most important data items in the record will require a little extra work, if, once extracted, you want to understand their meaning.
These are the morphology codes and the anatomic site codes.
The morphology code occupies bytes 53-57 (for ICDO-3) and bytes 48-52 (for ICDO-2), for each record.
Examples of morphology codes are:
M96783 primary effusion lymphoma
M83703 adrenal cortical carcinoma
You can download a copy of the ICDO-3 codes from SEER
If you start with a pdf file of the ICDO codes, you will need to cut and paste the pdf file into a plain ascii text file, free of formatting characters, to use it in your scripts. I put the list of codes and equivalent terms into a text file that I named "ICDO-3".
I often use the following short subroutine to put the codes, and their term equivalents, into a hash. When I need to convert a code into its term, I just call the hash value.
open (ICD, "c\:\\ftp\\icd03\.txt");
$line = " ";
while ($line ne "")
{
$line = <ICD>;
if ($line =~ /([0-9]{4})\/([0-9]{1}) +/o)
{
$code = $1 . $2;
$term = $';
$term =~ s/ *\n//o;
$term = lc($term);
$dictionary{$code} = $term;
}
}
close ICD;
This snippet of code will mean something to you when you look at the data format in SEER's ICDO-3 file.
The same can be done with an ICDO-2 file, and with the anatomic site codes (the primary site item, bytes 43-46).
Anatomic site codes and terms are available at:
http://www.ncri.ie/data.cgi/html/icdo2sites.shtml
An example of a site code and its term equivalent is:
C649 Kidney NOS*
*NOS means not otherwise specified
Everything we want to do with the SEER files involves parsing through the files, line (record) by line; and then pulling out the data we're interested in.
If you want to do creative data mining, you will need to learn a little computer programming.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
An overview of the many uses of biomedical information is available in my book,
Biomedical Informatics.
More information on cancer is available in my recently published book, Neoplasms.
© 2008 Jules Berman
In June, 2014, my book, entitled Rare Diseases and Orphan Drugs: Keys to Understanding and Treating the Common Diseases was published by Elsevier. The book builds the argument that our best chance of curing the common diseases will come from studying and curing the rare diseases.
I urge you to read more about my book. There's a generous preview of the book at the Google Books site. If you like the book, please request your librarian to purchase a copy of this book for your library or reading room.
tags: common disease, orphan disease, orphan drugs, rare disease, subsets of disease, disease genetics, genetics of complex disease, genetics of common diseases, neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
Monday, November 17, 2008
Using SEER Public Use Data: 4
This blog continutes a series on the SEER Public Use Data files.
In a prior blog, I showed how to do a global search through the SEER public use neoplasm incidence files, producing a list of every neoplastic entity captured in the approx 3.5 million SEER cases, and categorizing them by ethnicity, average age of occurrence, number of occurrences, and other data culled from the master data sets.
Reviewing the output, I found that the Hispanic and White populations had a greater number of occurrences of germ cell tumors (an uncommon type of tumor) than the African-American population.
I went to the SEER site and used SEER's public query engine to see if this observation could be verified.
The SEER query site is:
http://seer.cancer.gov/canques/index.html
All queries begin here. I looked for tumor in males, in testes, comparing Hispanic non-whites with African-Americans. A simple interface permits these selections.
The SEER interface produces a list of your input parameters.

The same interface produces a bar chart of your results:
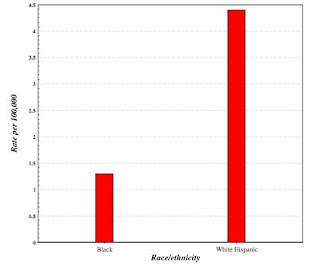
You may be wondering, if I am interested in germ cell tumors, why did I do a query on tumors of the testes. I did this because the SEER interface does not allow me to do a query on specific types of germ cell tumors or of any specific testicular tumor. I know that most testicular tumors are germ cell tumors, so I settled, figuring that if there were a difference in the incidence of germ cell tumors in the Hispanic and the African-American populations, it would show up in the query.
And that's what happened. The SEER output demonstrated that white Hispanics had a much higher incidence of testicular tumors (and, presumably, testicular germ cell tumors) than the African-American population.
If I want to find the ratio for specific tumors, I need to do a little more work. A simple Perl script produced the following list:
The left column is the ratio of cases per total population of white Hispanics divided by the same ratio for African-Americans. The second column is the number of cases, the third column is the average age of cases, and the final column is the ICD-0 term for the neoplasm.
We see (column 1) that white Hispanics have a higher case ratio for every type of germ cell tumor in males (regardless of site).
This tells us a few things. First, that all of the germ cell tumors are related to each other by more than histogenesis (cell of origin). They must have a relationship that extends to causation and development. Second, it tells us that the relatively high level of occurrence of germ cell tumors in white Hispanics is not just a fluke occurring in one cancer of one particular site. It is a consistent phenomenon that extends to several different related tumors and their histologic variants.
I should stress that germ cell tumors are rare, even in the Hispanic population. We are discussing relative rates of uncommon tumors among different ethnicities. An individual's risk of developing a germ cell tumor is low, regardless of ethnicity.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
More information on cancer is available in my recently published book, Neoplasms: Principles of Development and Diversity.
- © 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics
As requested in the SEER Public-Use Agreement, the SEER citation is included here:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
In a prior blog, I showed how to do a global search through the SEER public use neoplasm incidence files, producing a list of every neoplastic entity captured in the approx 3.5 million SEER cases, and categorizing them by ethnicity, average age of occurrence, number of occurrences, and other data culled from the master data sets.
Reviewing the output, I found that the Hispanic and White populations had a greater number of occurrences of germ cell tumors (an uncommon type of tumor) than the African-American population.
I went to the SEER site and used SEER's public query engine to see if this observation could be verified.
The SEER query site is:
http://seer.cancer.gov/canques/index.html
All queries begin here. I looked for tumor in males, in testes, comparing Hispanic non-whites with African-Americans. A simple interface permits these selections.
The SEER interface produces a list of your input parameters.

The same interface produces a bar chart of your results:

You may be wondering, if I am interested in germ cell tumors, why did I do a query on tumors of the testes. I did this because the SEER interface does not allow me to do a query on specific types of germ cell tumors or of any specific testicular tumor. I know that most testicular tumors are germ cell tumors, so I settled, figuring that if there were a difference in the incidence of germ cell tumors in the Hispanic and the African-American populations, it would show up in the query.
And that's what happened. The SEER output demonstrated that white Hispanics had a much higher incidence of testicular tumors (and, presumably, testicular germ cell tumors) than the African-American population.
If I want to find the ratio for specific tumors, I need to do a little more work. A simple Perl script produced the following list:
2.031 0173 026 germinoma
2.756 0005 038 intratubular malignant germ cells *
2.756 0005 025 malignant teratoma, undifferentiated type *
3.409 0104 019 teratoma, malignant nos
3.478 1125 035 seminoma nos
4.452 0054 036 seminoma, anaplastic type
4.757 0157 028 teratocarcinoma
5.053 0060 027 choriocarcinoma combined with teratoma
5.168 0018 050 spermatocytic seminoma *
5.523 0299 028 embryonal carcinoma nos
5.548 0363 028 mixed germ cell tumor
6.389 0028 027 germ cell tumor, nonseminomatous
* Cases with asterisks have too few cases (second column)
for any significance
The left column is the ratio of cases per total population of white Hispanics divided by the same ratio for African-Americans. The second column is the number of cases, the third column is the average age of cases, and the final column is the ICD-0 term for the neoplasm.
We see (column 1) that white Hispanics have a higher case ratio for every type of germ cell tumor in males (regardless of site).
This tells us a few things. First, that all of the germ cell tumors are related to each other by more than histogenesis (cell of origin). They must have a relationship that extends to causation and development. Second, it tells us that the relatively high level of occurrence of germ cell tumors in white Hispanics is not just a fluke occurring in one cancer of one particular site. It is a consistent phenomenon that extends to several different related tumors and their histologic variants.
I should stress that germ cell tumors are rare, even in the Hispanic population. We are discussing relative rates of uncommon tumors among different ethnicities. An individual's risk of developing a germ cell tumor is low, regardless of ethnicity.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
More information on cancer is available in my recently published book, Neoplasms: Principles of Development and Diversity.
- © 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics
As requested in the SEER Public-Use Agreement, the SEER citation is included here:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
Sunday, November 16, 2008
Using SEER Public Use Data: 3
Earlier today, I provided a blog with a large list of named neoplasms, listing ratios of occurrences in the African-American and white U.S. populations
http://julesberman.blogspot.com/2008/11/using-seer-public-use-data-2.html
For space limitions, the full name of the neoplasms was truncated.
I've prepared a separate web page, containing the full-length names of neoplasms, along with additional commentary.
http://www.julesberman.info/seerwhbl.htm
In June, 2014, my book, entitled Rare Diseases and Orphan Drugs: Keys to Understanding and Treating the Common Diseases was published by Elsevier. The book builds the argument that our best chance of curing the common diseases will come from studying and curing the rare diseases.
I urge you to read more about my book. There's a generous preview of the book at the Google Books site. If you like the book, please request your librarian to purchase a copy of this book for your library or reading room.
- Jules J. Berman, Ph.D., M.D.
http://julesberman.blogspot.com/2008/11/using-seer-public-use-data-2.html
For space limitions, the full name of the neoplasms was truncated.
I've prepared a separate web page, containing the full-length names of neoplasms, along with additional commentary.
http://www.julesberman.info/seerwhbl.htm
In June, 2014, my book, entitled Rare Diseases and Orphan Drugs: Keys to Understanding and Treating the Common Diseases was published by Elsevier. The book builds the argument that our best chance of curing the common diseases will come from studying and curing the rare diseases.
I urge you to read more about my book. There's a generous preview of the book at the Google Books site. If you like the book, please request your librarian to purchase a copy of this book for your library or reading room.
- Jules J. Berman, Ph.D., M.D.
Using SEER Public Use Data: 2
Continuing my previous blog on the SEER Public Use Data files, here is a listing of the ratios of cancer cases, of different types, occurring in the white population and the African-American population in the U.S.
As a rough guide to interpreting the data, neoplasms found in the top third of the list occur disproportionately often in the African-American population. Neoplasms found in the bottom third of the list occur disproportionately often in the white population. Neoplasms in the middle third occur in similar proportions in both sets.
The left hand column is the ratio of occurrence of the tumors in the two populations. Each ratio was calculated as the fraction of cases of the tumor in the white population divided by the fraction of the cases of the tumor in the black population. If the tumor accounted for the same fraction of total cancer cases in the white and black populations, it would have a ratio of 1. It is important to note that we cannot simply find the ratio of the the tumor's occurrence in the white and black populations (because there are many more whites than blacks).
You might be wondering why this question (i.e., tumors in blacks vs tumors in whites) has any clinical importance or biological relevance. It's a good question, and it doesn't have an answer that will satisfy everyone. First, the information helps us avoid diagnostic errors. For example, a pathologist should be wary about making a diagnosis of Ewing's tumor or of superficial spreading melanoma in an African American (in whom these tumors seldom occur). Second, the list generates new hypotheses. We notice that germ cell tumors (including seminomas, teratomas and embryonal carcinomas) occur much for frequently in whites than in the black population. Why is this? Is there some gene that contributes to these tumors, that occurs more frequenly in the white population? We cannot ask such questions if we do not have the kinds of observations included in this list. Third, the list can be generated with the Public Use data sets, which contain race/ethnicity data in case records.
In a later blog, I will show the short Perl script that generated the table from the SEER Public Use Data.
The first column is the case occurrence ratio (white/black). The second column is the number of cases, for each tumor type, in the SEER public use data sets. Tumors occurring with under 40 cases were excluded. The third column is the average age of patients with the neoplasm. The fourth column is the ICD-O(international classification of diseases - oncology) neoplasm term, truncated for space limitation.
In the next few blogs, I'll explain the clinical significance of this and other demo projects, and I'll describe the free, open source, techniques that I used to extract and compile the data. Afterwards, I'll show you how we can drill into the data to refine the questions we can ask, so that we can draw conclusions that can stand up to critical inspection.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
More information on cancer is available in my recently published book, Neoplasms.
- © 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
As a rough guide to interpreting the data, neoplasms found in the top third of the list occur disproportionately often in the African-American population. Neoplasms found in the bottom third of the list occur disproportionately often in the white population. Neoplasms in the middle third occur in similar proportions in both sets.
The left hand column is the ratio of occurrence of the tumors in the two populations. Each ratio was calculated as the fraction of cases of the tumor in the white population divided by the fraction of the cases of the tumor in the black population. If the tumor accounted for the same fraction of total cancer cases in the white and black populations, it would have a ratio of 1. It is important to note that we cannot simply find the ratio of the the tumor's occurrence in the white and black populations (because there are many more whites than blacks).
You might be wondering why this question (i.e., tumors in blacks vs tumors in whites) has any clinical importance or biological relevance. It's a good question, and it doesn't have an answer that will satisfy everyone. First, the information helps us avoid diagnostic errors. For example, a pathologist should be wary about making a diagnosis of Ewing's tumor or of superficial spreading melanoma in an African American (in whom these tumors seldom occur). Second, the list generates new hypotheses. We notice that germ cell tumors (including seminomas, teratomas and embryonal carcinomas) occur much for frequently in whites than in the black population. Why is this? Is there some gene that contributes to these tumors, that occurs more frequenly in the white population? We cannot ask such questions if we do not have the kinds of observations included in this list. Third, the list can be generated with the Public Use data sets, which contain race/ethnicity data in case records.
In a later blog, I will show the short Perl script that generated the table from the SEER Public Use Data.
The first column is the case occurrence ratio (white/black). The second column is the number of cases, for each tumor type, in the SEER public use data sets. Tumors occurring with under 40 cases were excluded. The third column is the average age of patients with the neoplasm. The fourth column is the ICD-O(international classification of diseases - oncology) neoplasm term, truncated for space limitation.
White/Black No. cases Avg age ICD-0 Diagnosis
00.113 0000040 040 pigmented dermatofibrosarcoma protuberans
00.190 0000126 060 adult t-cell leukemia/lymphoma (htlv-1 pos.)
00.225 0000056 068 granular cell tumor, malignant
00.227 0000057 061 collecting duct carcinoma
00.238 0000045 056 thymoma, type ab, malignant
00.263 0000089 050 ameloblastoma, malignant
00.271 0000053 053 hypereosinophilic syndrome
00.314 0000116 051 gastrinoma, malignant
00.314 0000042 053 odontogenic tumor, malignant
00.372 0000103 026 alveolar soft part sarcoma
00.394 0000120 055 atypical medullary carcinoma
00.405 0000335 051 medullary carcinoma with lymphoid stroma
00.415 0000053 038 craniopharyngioma
00.416 0000481 039 hodgkin lymph., nodular lymphocyte predom.
00.419 0000115 030 precursor t-cell lymphoblastic lymphoma
00.428 0000200 065 plasma cell leukemia
00.439 0000832 031 choriocarcinoma
00.441 0000050 002 infantile fibrosarcoma
00.443 0001263 062 gastrointestinal stromal sarcoma
00.444 0003634 042 dermatofibrosarcoma nos
00.447 0000040 040 sertoli-leydig cell tumor, poorly differentiated
00.448 0000106 038 mesenchymal chondrosarcoma
00.461 0001225 050 pituitary adenoma, nos
00.461 0000042 066 prolymphocytic leukemia, t-cell type
00.464 0000959 055 thymoma, malignant
00.484 0000091 059 polymorphous low grade adenocarcinoma
00.485 0019957 064 hepatocellular carcinoma nos
00.494 0000780 053 granulosa cell tumor, malignant
00.496 0000227 028 chondroblastic osteosarcoma
00.501 0000779 067 mesodermal mixed tumor
00.501 0023109 063 squamous cell carcinoma, keratinizing type nos
00.502 0002766 072 adenocarcinoma, intestinal type
00.504 0036429 069 multiple myeloma
00.507 0000641 001 retinoblastoma nos
00.510 0000482 045 sq. cell carcinoma, keratinizing, nos, in situ
00.511 0001466 005 nephroblastoma nos
00.514 0000140 056 pleomorphic rhabdomyosarcoma
00.515 0000303 061 metaplastic carcinoma, nos
00.517 0000117 024 precursor t-cell lymphoblastic leukemia
00.520 0000337 018 alveolar rhabdomyosarcoma
00.531 0000105 065 adenocarcinoma with neuroendocrine differen.
00.533 0000117 051 paraganglioma, malignant
00.541 0003377 060 mycosis fungoides
00.544 0003088 068 mullerian mixed tumor
00.550 0000756 014 embryonal rhabdomyosarcoma
00.551 0007534 053 medullary carcinoma nos
00.553 0003575 070 tumor cells, malignant
00.553 0000617 060 renal cell carcinoma, chromophobe type
00.553 0000299 060 squamous cell carcinoma, small cell, nonkeratining
00.554 0000514 067 intracystic carcinoma, nos
00.558 0000118 028 parosteal osteosarcoma
00.565 0000070 055 juvenile carcinoma of the breast
00.570 0057571 038 carcinoma in situ nos
00.571 0000196 049 pheochromocytoma, malignant
00.573 0000156 033 synovial sarcoma, biphasic type
00.577 0000054 058 malignant myoepithelioma
00.584 0000114 059 atypical meningioma
00.585 0000041 000 retinoblastoma, differentiated type
00.586 0000486 060 epithelioid leiomyosarcoma
00.596 0000183 065 superficial spreading adenocarcinoma
00.597 0001267 066 carcinoma, diffuse type
00.598 0000906 061 meningioma, malignant
00.602 0000191 058 stromal sarcoma, nos
00.613 0001094 065 intraductal papillary adenocarcinoma with invasion
00.617 0000135 063 composite carcinoid
00.626 0000043 002 retinoblastoma, undifferentiated type
00.627 0000118 037 giant cell tumor of bone, malignant
00.629 0001848 033 osteosarcoma nos
00.630 0016028 060 carcinoid tumor, malignant
00.631 0000090 024 malignant rhabdoid tumor
00.633 0000202 058 myeloid sarcoma
00.634 0000403 057 adenosarcoma
00.634 0000229 058 meningotheliomatous meningioma
00.637 0000757 064 papillary squamous cell carcinoma
00.637 0008089 058 squamous cell carcinoma, large cell, nonkeratinizing
00.638 0003873 044 squamous cell carcinoma, microinvasive
00.639 0000069 021 ganglioglioma
00.639 0000118 062 epithelial-myoepithelial carcinoma
00.641 0000586 069 chronic myeloproliferative disease, nos
00.643 0001258 052 fibrosarcoma nos
00.654 0000884 051 lymphoepithelial carcinoma
00.658 0014107 041 kaposi's sarcoma
00.661 0000690 047 neurofibrosarcoma
00.662 0000514 065 pleomorphic carcinoma
00.667 0247826 064 squamous cell carcinoma nos
00.673 0000082 059 thymic carcinoma, nos
00.673 0000047 047 leydig cell tumor, malignant
00.673 0000050 066 adenocarc. in situ in mult. adenomatous polyps
00.674 0000179 049 mesenchymoma, malignant
00.678 0014878 068 non-small cell carcinoma
00.681 0000890 054 anaplastic large cell lymphoma, t-cell and null cell
00.682 0002723 065 meningioma nos
00.682 0000861 042 hodgkin's disease, lymphocytic predominance
00.683 0000114 033 fibroblastic osteosarcoma
00.685 0000282 042 epithelioid cell sarcoma
00.685 0000507 019 primitive neuroectodermal tumor
00.685 0000123 040 clear cell sarcoma of tendons and aponeuroses
00.686 0000087 037 small cell sarcoma
00.686 0000334 063 giant cell sarcoma (except of bone m9250/3)
00.687 0008070 059 leiomyosarcoma nos
00.688 0003285 057 inflammatory carcinoma
00.690 0000555 037 synovial sarcoma nos
00.692 0000058 052 hodgkin's disease, lymphocytic depletion, diffuse fib
00.693 0001762 004 neuroblastoma nos
00.693 0000257 048 megakaryocytic leukemia
00.697 0000939 063 plasmacytoma, extramedullary
00.702 0003124 059 sarcoma nos
00.703 0004558 059 infiltr. duct mixed with other types of carcinoma, in
00.706 0002264 068 carcinosarcoma nos
00.707 0001065 064 giant cell carcinoma
00.707 0000053 026 telangiectatic osteosarcoma
00.707 0000084 014 langerhans cell histiocytosis, disseminated
00.712 0002593 062 infiltr. duct mixed with other types of carcinoma
00.727 0000049 022 precursor b-cell lymphoblastic lymphoma
00.728 0010967 065 signet ring cell carcinoma
00.735 0000723 064 apocrine adenocarcinoma
00.745 0000903 055 endometrial stromal sarcoma
00.750 0001267 060 mature t-cell lymphoma, nos
00.751 0000085 050 acute myeloid leukemia, t(8;21)(q22;q22)
00.755 0000869 063 plasma cell tumor, malignant
00.755 0000194 048 endometrial stromal sarcoma, low grade
00.761 0010254 064 carcinoma, undifferentiated type nos
00.762 0003162 062 noninfiltrating intraductal papillary adenocarcinoma
00.763 0000934 052 cystosarcoma phyllodes, malignant
00.768 0000295 068 pseudosarcomatous carcinoma
00.773 0011520 064 acinar cell carcinoma
00.774 0000584 067 squamous cell carcinoma, spindle cell type
00.782 0000493 062 basaloid squamous cell carcinoma
00.785 0029592 065 large cell carcinoma nos
00.786 0000787 066 spindle cell carcinoma
00.786 0000277 063 combined hepatocellular carcinoma and cholangiocarcino
00.791 0025451 067 mucin-producing adenocarcinoma
00.794 0001025 060 spindle cell sarcoma
00.796 0006891 056 comedocarcinoma nos
00.796 0000258 062 renal cell carcinoma, sarcomatoid
00.800 0000123 065 brenner tumor, malignant
00.805 0016555 068 adenocarcinoma in tubulovillous adenoma
00.808 0000051 035 prolactinoma
00.809 0005831 067 adenocarcinoma in situ in tubulovillous adenoma
00.811 0024527 041 squamous intraepithelial neoplasia, grade iii
00.812 0000234 058 malignant tumor, small cell type
00.812 0000248 052 chronic myelogenous leukemia, bcr/abl positive
00.814 0004050 050 follicular adenocarcinoma nos
00.824 0000856 065 adenocarcinoma with mixed subtypes
00.824 0000060 016 choroid plexus papilloma, malignant
00.828 0000046 053 fascial fibrosarcoma
00.829 0000843 058 intraductal micropapillary carcinoma
00.831 0000947 064 granular cell carcinoma
00.833 0000079 044 papillary cystadenoma, borderline malignancy (c56.9)
00.839 0000324 061 acute myeloid leukemia, minimal differentiation
00.841 0000584 020 endodermal sinus tumor
00.845 0000722 050 neurilemmoma, malignant
00.845 0000557 067 combined small cell carcinoma
00.846 0002217 061 paget's disease and infiltrating duct carcinoma of brea
00.851 0000505 043 rhabdomyosarcoma nos
00.851 0011193 063 adenosquamous carcinoma
00.852 0000265 053 fibromyxosarcoma
00.854 0000139 022 ependymoma, anaplastic type
00.858 0000066 057 thymoma, type b1, malignant
00.858 0000045 037 mediastinal large b-cell lymphoma
00.858 0000388 037 sq. cell carcinoma, lg. cell, non-ker., in situ
00.859 0000416 038 hodgkin lymphoma, nod. scler., grade 1
00.861 0000517 047 follicular adenocarcinoma, well differentiated type
00.868 0000052 065 eccrine adenocarcinoma
00.868 0063370 038 squamous cell carcinoma in situ nos
00.870 0037543 064 renal cell carcinoma
00.871 0001654 066 linitis plastica
00.873 0001021 060 liposarcoma, well differentiated type
00.874 0000328 055 adenocarcinoid tumor
00.874 0000382 061 mixed tumor, malignant nos
00.877 0002754 056 intraductal and lobular in situ carcinoma
00.877 0000141 050 follicular adenocarcinoma, trabecular type
00.878 0000100 061 papillary squamous cell carcinoma, non-invasive
00.880 0001242 023 teratoma, malignant nos
00.880 0000080 071 refract. anemia with excess blasts in transformation
00.883 0005106 064 neuroendocrine carcinoma
00.884 0182616 072 carcinoma nos
00.884 0003757 054 mucoepidermoid carcinoma
00.885 0000179 051 adenocarcinoma in adenomatous polyposis coli
00.897 0000100 019 desmoplastic medulloblastoma
00.900 0000690 028 germinoma
00.904 1021940 068 adenocarcinoma nos
00.907 0056558 076 neoplasm, malignant
00.909 0000055 020 clear cell sarcoma of kidney
00.920 0003319 058 adenoid cystic carcinoma
00.921 0000082 032 neuroepithelioma nos
00.923 0010280 061 chronic myeloid leukemia
00.927 0003078 048 hodgkin's disease nos
00.927 0000963 035 precursor cell lymphoblastic lymphoma, nos
00.930 0000165 067 bronchiolo-alveolar carcinoma, non-mucinous
00.933 0000884 062 acral lentiginous melanoma, malig.
00.934 0000308 007 ganglioneuroblastoma
00.937 0000265 055 mucocarcinoid tumor, malignant
00.937 0000403 066 large cell neuroendocrine carcinoma
00.946 0000089 057 myxosarcoma
00.958 0000820 073 refractory anemia
00.959 0013199 064 malignant lymphoma nos
00.963 0000315 066 malignant tumor, fusiform cell type
00.966 0000086 045 hemangioblastoma
00.967 0000855 057 islet cell carcinoma
00.968 0001163 013 medulloblastoma nos
00.968 0000191 052 myxoid chondrosarcoma
00.970 0001140 047 acute promyelocytic leukemia
00.972 0000096 056 myxoid leiomyosarcoma
00.972 0000093 025 pleomorphic xanthoastrocytoma
00.972 0000087 064 acute panmyelosis with myelofibrosis
00.974 0002700 050 bowen's disease
00.981 0000080 063 basal cell adenocarcinoma
00.982 0000125 065 nodular hidradenoma, malignant
00.989 0000114 067 sezary's disease
00.989 0000115 051 round cell liposarcoma
00.992 0035630 059 intraductal carcinoma, noninfiltrating nos
00.993 0000358 051 hemangiopericytoma, malignant
00.996 0003164 064 scirrhous adenocarcinoma
00.997 0003337 046 glioma, malignant
01.003 0001045 060 cutaneous t-cell lymphoma, nos
01.027 0001609 037 burkitt lymphoma, nos
01.028 0001015 067 lymphoid leukemia nos
01.032 0000104 048 epithelioid hemangioendothelioma, malignant
01.036 0000303 004 hepatoblastoma
01.036 0000998 065 essential thrombocythemia
01.040 0000390 066 alveolar adenocarcinoma
01.047 0000094 035 protoplasmic astrocytoma
01.050 0016754 069 adenocarcinoma in villous adenoma
01.050 0001041 049 serous cystadenoma, borderline malignancy (c56.9)
01.058 0000548 038 hodgkin lymphoma, nod. scler., cellular phase
01.059 0001545 017 pilocytic astrocytoma (c71._) 9421/1
01.061 0008163 067 adenocarcinoma in situ in adenomatous polyp
01.065 0000815 066 erythroleukemia
01.066 0001669 066 small cell carcinoma, intermediate cell
01.067 0000248 064 giant cell and spindle cell carcinoma
01.071 0001388 034 ependymoma nos
01.072 0000624 066 papillary carcinoma in situ
01.075 0004414 046 hodgkin's disease, mixed cellularity
01.089 0001323 062 liposarcoma nos
01.092 0000142 054 mesonephroma, malignant
01.092 0000212 067 bronchiolo-alveolar carcinoma, mucinous
01.093 0001167 053 myxoid liposarcoma
01.100 0050173 066 small cell carcinoma nos
01.103 0004446 065 carcinoma, anaplastic type nos
01.111 0000159 052 hemangioendothelioma, malignant
01.116 0002422 074 myelodysplastic syndrome, nos
01.117 0333623 061 infiltrating duct carcinoma
01.120 0000405 064 pleomorphic liposarcoma
01.132 0005812 062 fibrous histiocytoma, malignant
01.132 0003638 065 marginal zone b-cell lymphoma, nos
01.134 0004672 069 adenocarcinoma in situ in villous adenoma
01.138 0000145 057 mixed type liposarcoma
01.144 0000084 063 psammomatous meningioma
01.149 0000166 060 carcinoid tumor, argentaffin, malignant
01.154 0000096 039 hepatocellular carcinoma, fibrolamellar
01.162 0000655 074 refractory anemia with sideroblasts
01.166 0000151 064 adenocarcinoma in mult. adenomatous polyps
01.166 0001603 048 serous papillary cystic tumor of borderline malignancy
01.169 0011933 033 hodgkin lymphoma, nodular sclerosis, nos
01.171 0018131 061 acute myeloid leukemia
01.173 0000287 063 dedifferentiated liposarcoma
01.173 0009679 058 comedocarcinoma, noninfiltrating
01.175 0027965 059 papillary adenocarcinoma nos
01.178 0000091 045 papillary carcinoma, encapsulated
01.184 0001275 064 polycythemia vera
01.189 0020880 068 adenocarcinoma in adenomatous polyp
01.189 0000827 021 precursor b-cell lymphoblastic leukemia
01.190 0000215 046 follicular carcinoma, minimally invasive
01.191 0000461 066 basaloid carcinoma
01.196 0000178 037 synovial sarcoma, spindle cell type
01.196 0000362 069 myelosclerosis with myeloid metaplasia
01.197 0001872 069 myeloid leukemia nos
01.198 0045778 068 mucinous adenocarcinoma
01.200 0002839 059 cribriform carcinoma in situ
01.206 0000516 050 papillary microcarcinoma
01.208 0000435 036 hodgkin lymphoma, nod. scler., grade 2
01.220 0000165 061 carcinoma in pleomorphic adenoma
01.223 0000614 060 acute myeloid leukemia with maturation
01.234 0001092 062 mixed cell adenocarcinoma
01.243 0009222 023 precursor cell lymphoblastic leukemia, nos
01.249 0020557 061 clear cell adenocarcinoma nos
01.254 0011881 066 bronchiolo-alveolar adenocarcinoma
01.255 0000115 044 adenocarcinoma, endocervical type
01.257 0001270 066 ml, lymphoplasmacytic
01.262 0000144 056 transitional meningioma
01.263 0004480 058 ml, large b-cell, diffuse, immunoblastic, nos
01.265 0010260 046 papillary and follicular adenocarcinoma
01.279 0000172 050 giant cell glioblastoma
01.287 0000057 055 chromophobe carcinoma
01.287 0000127 066 neoplasm, uncertain whether benign or malignant
01.288 0001435 058 oxyphilic adenocarcinoma
01.295 0000880 065 cribriform carcinoma
01.300 0000485 056 papillary mucinous cystadenocarcinoma
01.302 0000149 034 peripheral neuroectodermal tumor
01.312 0004610 069 cholangiocarcinoma
01.312 0002006 061 acute myelomonocytic leukemia
01.313 0000626 056 hodgkin's disease, lymphocytic depletion nos
01.320 0015063 062 papillary serous cystadenocarcinoma
01.326 0000241 066 angioimmunoblastic t-cell lymphoma
01.329 0000117 054 nk/t-cell lymphoma, nasal and nasal-type
01.336 0000758 068 villous adenocarcinoma
01.341 0000761 051 adrenal cortical carcinoma
01.348 0001142 063 cystadenocarcinoma nos
01.356 0000618 066 paget's disease, mammary
01.362 0009635 067 ml, small b lymphocytic, nos
01.363 0003731 065 acute leukemia nos
01.364 0001353 063 hemangiosarcoma
01.371 0008335 047 astrocytoma nos
01.372 0001358 064 cloacogenic carcinoma
01.376 0010414 054 lobular carcinoma in situ
01.391 0021695 061 infiltrating duct and lobular carcinoma
01.397 0017866 064 oat cell carcinoma
01.407 0002708 063 serous surface papillary carcinoma
01.414 0000054 065 hepatocellular carcinoma, clear cell type
01.425 0000285 046 burkitt's tumor
01.425 0003428 056 mucinous cystadenocarcinoma nos
01.437 0004682 062 serous cystadenocarcinoma nos
01.445 0000301 070 prolymphocytic leukemia, nos
01.451 0000327 067 acute myeloid leuk. with multilineage dysplasia
01.454 0000156 069 basosquamous carcinoma
01.455 0037088 063 ml, large b-cell, diffuse
01.481 0000050 059 solitary fibrous tumor, malignant
01.487 0001358 063 paget disease and intraductal ca.
01.487 0001004 050 medullary carcinoma with amyloid stroma
01.488 0012432 062 malignant lymphoma, non hodgkin's type
01.491 0004319 065 ml, mixed sm. and lg. cell, diffuse
01.497 0001673 068 verrucous carcinoma nos
01.512 0002581 072 leukemia nos
01.522 0001770 058 acute monocytic leukemia
01.530 0001669 060 duct carcinoma in situ, solid type
01.535 0000086 065 adenoid squamous cell carcinoma
01.535 0000086 062 therapy-related myelodysplastic syndrome, nos
01.540 0000079 058 thymoma, type b3, malignant
01.559 0030328 070 chronic lymphoid leukemia
01.599 0000111 047 papillary mucinous cystadenoma, borderline malignancy (
01.605 0000179 067 splenic marginal zone b-cell lymphoma
01.617 0001798 074 chronic myelomonocytic leukemia, nos
01.627 0006684 058 adenocarcinoma in situ
01.632 0000111 055 fibrous meningioma
01.641 0000074 061 hodgkin's disease, lymphocytic depletion, reticular
01.650 0000763 072 refractory anemia with excess blasts
01.651 0022382 050 papillary carcinoma nos
01.658 0000885 061 infiltrating ductular carcinoma
01.666 0000118 042 burkitt cell leukemia
01.692 0001301 059 papillary cystadenocarcinoma nos
01.721 0050243 070 transitional cell carcinoma nos
01.733 0002320 048 astrocytoma, anaplastic type
01.738 0000363 064 sweat gland adenocarcinoma
01.742 0026705 061 endometrioid carcinoma
01.777 0000500 048 oligodendroglioma, anaplastic type
01.796 0034336 064 lobular carcinoma nos
01.818 0000295 061 glioblastoma with sarcomatous component
01.829 0004897 068 mesothelioma, malignant
01.846 0000302 052 esthesioneuroblastoma
01.863 0001932 051 chondrosarcoma nos
01.868 0000042 061 adenocarcinoma with apocrine metaplasia
01.905 0000472 063 infiltrating lobular mixed with other types of carc.
01.924 0000448 059 acute myeloid leukemia without maturation
01.931 0001012 040 mixed glioma
01.935 0000159 049 nonencapsulated sclerosing carcinoma
01.986 0000128 053 spermatocytic seminoma
01.987 0001486 049 mucinous cystic tumor of borderline malignancy (c56.9)
01.994 0002890 066 mantle cell lymphoma
02.002 0000764 042 fibrillary astrocytoma
02.020 0000184 067 trabecular adenocarcinoma
02.020 0000046 069 eccrine poroma, malignant
02.034 0003345 063 malignant lymphoma, nodular nos
02.046 0000966 031 dysgerminoma
02.053 0000070 040 myxopapillary ependymoma
02.057 0006337 062 tubular adenocarcinoma
02.088 0000978 052 neurilemmoma nos
02.090 0000219 038 astroblastoma
02.119 0003414 062 malignant lymphoma, follicular center cell, noncleaved,
02.171 0019907 061 glioblastoma nos
02.222 0000239 063 solid carcinoma nos
02.229 0001585 071 waldenstrom macroglobulinemia
02.235 0000940 069 transitional cell carcinoma in situ
02.242 0000122 071 transitional cell carcinoma, spindle cell type
02.289 0000077 062 atypical carcinoid tumor
02.333 0002297 041 oligodendroglioma nos
02.363 0000263 071 refractory cytopenia with multilineage dysplasia
02.491 0000089 074 myelodysplastic syndr. with 5q deletion syndrome
02.525 0000054 068 carcinoma simplex
02.554 0000461 049 gemistocytic astrocytoma
02.558 0000083 064 small cell carcinoma, fusiform cell type
02.592 0000086 056 papillary carcinoma, columnar cell
02.657 0000408 072 klatskin tumor
02.666 0000147 056 primary cutan. cd30+ t-cell lymphoprolif. disorder
02.726 0096537 068 papillary transitional cell carcinoma
02.790 0005812 062 malignant lymphoma, mixed lymphocytic-histiocytic, nod
02.891 0000614 055 chordoma
02.932 0008938 060 malignant lymphoma, follicular center cell, cleaved,
02.962 0000476 069 fibrous mesothelioma, malignant
03.094 0002388 030 mixed germ cell tumor
03.131 0000786 072 basal cell carcinoma nos
03.245 0002438 069 papillary trans. cell carcinoma, non-invasive
03.257 0000135 054 malignant mastocytosis
03.282 0000494 062 skin appendage carcinoma
03.491 0000261 072 mesothelioma, biphasic type, malignant
03.570 0000796 072 sebaceous adenocarcinoma
03.587 0001263 067 epithelioid mesothelioma, malignant
03.602 0000113 060 queyrat's erythroplasia
03.602 0000116 065 sclerosing sweat duct carcinoma
03.694 0002147 058 hairy cell leukemia
03.695 0004086 059 adenocarcinoma with squamous metaplasia
03.737 0000042 056 gliomatosis cerebri
04.040 0000041 064 lymphangiosarcoma
04.040 0000095 028 germ cell tumor, nonseminomatous
04.337 0009849 037 seminoma nos
04.747 0000049 070 osteosarcoma in paget's disease of bone
04.955 0001717 028 teratocarcinoma
05.050 0000057 048 ac. myelomonocytic leuk. w abn. mar. eosinophils
05.529 0003184 030 embryonal carcinoma nos
05.611 0001088 019 ewing's sarcoma
05.824 0000556 028 choriocarcinoma combined with teratoma
06.019 0000627 038 seminoma, anaplastic type
06.565 0000401 060 epithelioid cell melanoma
08.055 0000811 071 paget's disease, extramammary (except paget's of bone)
08.206 0001716 074 merkel cell carcinoma
10.024 0000412 056 malignant melanoma, regressing
10.706 0000110 052 precancerous melanosis nos
10.891 0000661 061 mixed epithel. & spindle cell melanoma
12.623 0048315 056 malignant melanoma nos
18.888 0000764 060 amelanotic melanoma
19.367 0000783 066 desmoplastic melanoma, malignant
21.009 0001281 062 spindle cell melanoma nos
24.492 0008688 060 nodular melanoma
29.788 0007272 069 malignant melanoma in hutchinson's melanotic freckle
31.608 0021999 057 melanoma in situ
38.463 0040929 052 superficial spreading melanoma
38.730 0018652 068 hutchinson's melanotic freckle
47.472 0000477 058 spindle cell melanoma, type b
62.046 0004547 051 superficial spreading melanoma, in situ
In the next few blogs, I'll explain the clinical significance of this and other demo projects, and I'll describe the free, open source, techniques that I used to extract and compile the data. Afterwards, I'll show you how we can drill into the data to refine the questions we can ask, so that we can draw conclusions that can stand up to critical inspection.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
More information on cancer is available in my recently published book, Neoplasms.
- © 2008 Jules Berman
key words: neoplasms, cancer, neoplasia, precancer, tumor, tumour, tumors, tumours, neoplasm, carcinogenesis, carcinogens, tumor genetics
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
Friday, November 14, 2008
Using SEER Public Use Data: 1
SEER is the U.S. National Cancer Institute's Surveillance, Epidemiology and End Results program. It is an amazing resource for information about the cancers that occur in the U.S. One of the products of SEER is the Public Use dataset, which contains de-identified records on over 3.5 million cancers that have occurred between 1973 and 2005.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
I thought I would do a series of blogs, extended over the next few weeks or months, showing how the SEER dataset can be analyzed, the kinds of hypotheses and discoveries that can be made by studying the database, and the kinds of things that you can do when you combine SEER data with data from other publicly available resources.
Each SEER record is a cancer case, described by a series of 258 (mostly) numbers, in byte-assigned positions, described by a data dictionary document. When you have the byte locations for the data dictionary entries, you can easily write a short script (I like to use Perl, Ruby, or Python) that can extract and compile data any way you wish.
For example, the following list is a compilation of all of the diagnoses (that occur at least 10 times in the data set), sorted by the age of the person at the time of diagnosis, accompanied by the number of cases and the cumulative fraction of cases accounted for, and by the diagnosis (truncated for reasons of space).
As specified in the Limited-Use Data Agreement, the citation for the SEER data
is as follows:
Once we have the columned data, we can easily produce a graphic that represents the salient features we would like to emphasize.

In the next few blogs, I'll explain the clinical significance of this little demo project, and I'll describe the free, open source, techniques that I used to extract and compile the data. Afterwards, I'll show you how we can drill into the data to refine the questions we can ask, so that we can draw conclusions that can stand up to critical inspection.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
- © 2008 Jules Berman
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
In June, 2014, my book, entitled Rare Diseases and Orphan Drugs: Keys to Understanding and Treating the Common Diseases was published by Elsevier. The book builds the argument that our best chance of curing the common diseases will come from studying and curing the rare diseases.
I urge you to read more about my book. There's a generous preview of the book at the Google Books site. If you like the book, please request your librarian to purchase a copy of this book for your library or reading room.
When you have 3.5 million cancer cases to study, you can draw certain types of inferences that could not possibly be made with the data accumulated at a single medical institution.
I thought I would do a series of blogs, extended over the next few weeks or months, showing how the SEER dataset can be analyzed, the kinds of hypotheses and discoveries that can be made by studying the database, and the kinds of things that you can do when you combine SEER data with data from other publicly available resources.
Each SEER record is a cancer case, described by a series of 258 (mostly) numbers, in byte-assigned positions, described by a data dictionary document. When you have the byte locations for the data dictionary entries, you can easily write a short script (I like to use Perl, Ruby, or Python) that can extract and compile data any way you wish.
For example, the following list is a compilation of all of the diagnoses (that occur at least 10 times in the data set), sorted by the age of the person at the time of diagnosis, accompanied by the number of cases and the cumulative fraction of cases accounted for, and by the diagnosis (truncated for reasons of space).
Age Number Cumu- Name
yrs of lat- of
cases ive Neoplasm
-------------------------------------------------
000 0000041 0.000 retinoblastoma, differentiated
001 0000641 0.000 retinoblastoma nos
002 0000050 0.000 infantile fibrosarcoma
002 0000016 0.000 juvenile myelomonocytic leukemi
002 0000021 0.000 atypical teratoid/rhabdoid tumo
002 0000043 0.000 retinoblastoma, undifferentiate
004 0000303 0.000 hepatoblastoma
004 0001762 0.000 neuroblastoma nos
004 0000012 0.000 medulloepithelioma nos
005 0001466 0.001 nephroblastoma nos
007 0000308 0.001 ganglioneuroblastoma
011 0000013 0.001 subependymal giant cell astrocy
012 0000010 0.001 pancreatoblastoma
013 0001163 0.001 medulloblastoma nos
013 0000018 0.001 neurofibromatosis nos
014 0000756 0.001 embryonal rhabdomyosarcoma
014 0000084 0.001 langerhans cell histiocytosis,
016 0000060 0.001 choroid plexus papilloma, malig
017 0001545 0.002 pilocytic astrocytoma (c71._) 9
018 0000031 0.002 embryonal sarcoma
018 0000337 0.002 alveolar rhabdomyosarcoma
019 0001088 0.002 ewing's sarcoma
019 0000100 0.002 desmoplastic medulloblastoma
019 0000507 0.002 primitive neuroectodermal tumor
020 0000584 0.003 endodermal sinus tumor
020 0000055 0.003 clear cell sarcoma of kidney
021 0000069 0.003 ganglioglioma
021 0000827 0.003 precursor b-cell lymphoblastic
022 0000139 0.003 ependymoma, anaplastic type
022 0000025 0.003 dysembryoplastic neuroepithelia
022 0000049 0.003 precursor b-cell lymphoblastic
023 0001242 0.003 teratoma, malignant nos
023 0000021 0.003 choroid plexus papilloma nos
023 0009222 0.006 precursor cell lymphoblastic le
024 0000098 0.006 pineoblastoma
024 0000090 0.006 malignant rhabdoid tumor
024 0000117 0.006 precursor t-cell lymphoblastic
025 0000011 0.006 periosteal osteosarcoma
025 0000023 0.006 mixed type rhabdomyosarcoma
025 0000093 0.006 pleomorphic xanthoastrocytoma
026 0000103 0.006 alveolar soft part sarcoma
026 0000019 0.006 chondroblastoma, malignant
026 0000053 0.006 telangiectatic osteosarcoma
027 0000011 0.006 spindle cell rhabdomyosarcoma
027 0000039 0.006 desmoplastic small round cell t
028 0000690 0.006 germinoma
028 0001717 0.007 teratocarcinoma
028 0000118 0.007 parosteal osteosarcoma
028 0000227 0.007 chondroblastic osteosarcoma
028 0000095 0.007 germ cell tumor, nonseminomatou
028 0000556 0.007 choriocarcinoma combined with t
029 0000023 0.007 ganglioglioma, anaplastic
029 0000019 0.007 intratubular malignant germ cel
029 0000021 0.007 malignant placental site tropho
030 0002388 0.008 mixed germ cell tumor
030 0003184 0.009 embryonal carcinoma nos
030 0000115 0.009 precursor t-cell lymphoblastic
031 0000966 0.009 dysgerminoma
031 0000832 0.009 choriocarcinoma
031 0000021 0.009 centrol neurocytoma
032 0000016 0.009 lipoma nos
032 0000082 0.009 neuroepithelioma nos
032 0000027 0.009 adamantinomatous craniopharyngi
032 0000019 0.009 malignant teratoma, undifferent
033 0001848 0.010 osteosarcoma nos
033 0000114 0.010 fibroblastic osteosarcoma
033 0000030 0.010 adamantinoma of long bones
033 0000156 0.010 synovial sarcoma, biphasic type
033 0011933 0.013 hodgkin lymphoma, nodular scler
034 0001388 0.014 ependymoma nos
034 0000149 0.014 peripheral neuroectodermal tumo
035 0000051 0.014 prolactinoma
035 0000094 0.014 protoplasmic astrocytoma
035 0000963 0.014 precursor cell lymphoblastic ly
036 0000435 0.014 hodgkin lymphoma, nod. scler.,
037 0009849 0.017 seminoma nos
037 0000087 0.017 small cell sarcoma
037 0000010 0.017 acidophil carcinoma
037 0000555 0.017 synovial sarcoma nos
037 0001609 0.017 burkitt lymphoma, nos
037 0000045 0.017 mediastinal large b-cell lympho
037 0000178 0.017 synovial sarcoma, spindle cell
037 0000118 0.018 giant cell tumor of bone, malig
037 0000388 0.018 sq. cell carcinoma, lg. cell, n
038 0000219 0.018 astroblastoma
038 0000053 0.018 craniopharyngioma
038 0057571 0.034 carcinoma in situ nos
038 0000016 0.034 androblastoma, malignant
038 0000627 0.034 seminoma, anaplastic type
038 0000106 0.034 mesenchymal chondrosarcoma
038 0063370 0.052 squamous cell carcinoma in situ
038 0000416 0.052 hodgkin lymphoma, nod. scler.,
038 0000548 0.052 hodgkin lymphoma, nod. scler.,
039 0000019 0.052 neurofibroma nos
039 0000026 0.052 sertoli cell carcinoma
039 0000096 0.052 hepatocellular carcinoma, fibro
039 0000021 0.052 hepatosplenic gamma-delta cell
039 0000481 0.052 hodgkin lymph., nodular lymphoc
039 0000024 0.052 mpnst with rhabdomyoblastic dif
040 0001012 0.053 mixed glioma
040 0000038 0.053 cavernous hemangioma
040 0000070 0.053 myxopapillary ependymoma
040 0000040 0.053 pigmented dermatofibrosarcoma p
040 0000123 0.053 clear cell sarcoma of tendons a
040 0000040 0.053 sertoli-leydig cell tumor, poor
041 0014107 0.057 kaposi's sarcoma
041 0002297 0.057 oligodendroglioma nos
041 0000014 0.057 spongioblastoma polare
041 0000010 0.057 papillary carcinoma, oxyphilic
041 0024527 0.064 squamous intraepithelial neopla
042 0000024 0.064 pulmonary blastoma
042 0000118 0.064 burkitt cell leukemia
042 0000764 0.065 fibrillary astrocytoma
042 0003634 0.066 dermatofibrosarcoma nos
042 0000282 0.066 epithelioid cell sarcoma
042 0000861 0.066 hodgkin's disease, lymphocytic
043 0000036 0.066 hemangioma nos
043 0000505 0.066 rhabdomyosarcoma nos
043 0000019 0.066 solid pseudopapillary carcinoma
044 0000015 0.066 papillary meningioma
044 0000013 0.066 papillary meningioma 9538/3
044 0000017 0.066 juxtacortical chondrosarcoma
044 0000115 0.066 adenocarcinoma, endocervical ty
044 0003873 0.067 squamous cell carcinoma, microi
044 0000079 0.067 papillary cystadenoma, borderli
045 0000086 0.067 hemangioblastoma
045 0000037 0.067 oligodendroblastoma
045 0000016 0.067 struma ovarii, malignant
045 0000029 0.067 undifferentiated sarcoma
045 0000010 0.067 clear cell chondrosarcoma
045 0000011 0.067 carotid body tumor, malignant
045 0000091 0.067 papillary carcinoma, encapsulat
045 0000035 0.067 extra-adrenal paraganglioma, ma
045 0000482 0.067 sq. cell carcinoma, keratinizin
046 0000285 0.068 burkitt's tumor
046 0003337 0.069 glioma, malignant
046 0000024 0.069 periosteal fibrosarcoma
046 0004414 0.070 hodgkin's disease, mixed cellul
046 0010260 0.073 papillary and follicular adenoc
046 0000215 0.073 follicular carcinoma, minimally
047 0008335 0.075 astrocytoma nos
047 0000690 0.075 neurofibrosarcoma
047 0000047 0.075 leydig cell tumor, malignant
047 0001140 0.076 acute promyelocytic leukemia
047 0000163 0.076 malignant melanoma in giant pig
047 0000517 0.076 follicular adenocarcinoma, well
047 0000111 0.076 papillary mucinous cystadenoma,
047 0000084 0.076 squamous cell carcinoma in situ
048 0003078 0.077 hodgkin's disease nos
048 0000257 0.077 megakaryocytic leukemia
048 0000039 0.077 ovarian stromal tumor, mal.
048 0002320 0.077 astrocytoma, anaplastic type
048 0000500 0.078 oligodendroglioma, anaplastic t
048 0000194 0.078 endometrial stromal sarcoma, lo
048 0000104 0.078 epithelioid hemangioendotheliom
048 0000057 0.078 ac. myelomonocytic leuk. w abn.
048 0001603 0.078 serous papillary cystic tumor o
049 0000033 0.078 subependymal glioma
049 0000179 0.078 mesenchymoma, malignant
049 0000461 0.078 gemistocytic astrocytoma
049 0000012 0.078 primary effusion lymphoma
049 0000196 0.078 pheochromocytoma, malignant
049 0000159 0.078 nonencapsulated sclerosing carc
049 0000034 0.078 dermoid cyst with malignant tra
049 0001041 0.079 serous cystadenoma, borderline
049 0001486 0.079 mucinous cystic tumor of border
050 0002700 0.080 bowen's disease
050 0000036 0.080 hodgkin's granuloma
050 0000012 0.080 chromophobe adenoma
050 0001225 0.080 pituitary adenoma, nos
050 0000722 0.080 neurilemmoma, malignant
050 0000172 0.081 giant cell glioblastoma
050 0022382 0.087 papillary carcinoma nos
050 0000089 0.087 ameloblastoma, malignant
050 0000516 0.087 papillary microcarcinoma
050 0000034 0.087 acute biphenotypic leukemia
050 0004050 0.088 follicular adenocarcinoma nos
050 0000017 0.088 mixed medullary-papillary carci
050 0001004 0.088 medullary carcinoma with amyloi
050 0000085 0.088 acute myeloid leukemia, t(8;21)
050 0000030 0.088 mucinous cystadenocarcinoma, no
050 0000029 0.088 mucinous adenocarcinoma, endoce
050 0000141 0.089 follicular adenocarcinoma, trab
051 0001932 0.089 chondrosarcoma nos
051 0000116 0.089 gastrinoma, malignant
051 0000115 0.089 round cell liposarcoma
051 0000117 0.089 paraganglioma, malignant
051 0000761 0.089 adrenal cortical carcinoma
051 0000884 0.090 lymphoepithelial carcinoma
051 0000358 0.090 hemangiopericytoma, malignant
051 0004547 0.091 superficial spreading melanoma,
051 0000335 0.091 medullary carcinoma with lympho
051 0000026 0.091 malignant giant cell tumor of s
051 0000179 0.091 adenocarcinoma in adenomatous p
052 0000043 0.091 myosarcoma
052 0000153 0.091 adenoma nos
052 0000978 0.091 neurilemmoma nos
052 0001258 0.092 fibrosarcoma nos
052 0000032 0.092 histiocytic sarcoma
052 0000191 0.092 myxoid chondrosarcoma
052 0000302 0.092 esthesioneuroblastoma
052 0000110 0.092 precancerous melanosis nos
052 0040929 0.104 superficial spreading melanoma
052 0000159 0.104 hemangioendothelioma, malignant
052 0000934 0.104 cystosarcoma phyllodes, maligna
052 0000031 0.104 malignant melanoma in precancer
052 0000248 0.104 chronic myelogenous leukemia, b
052 0000058 0.104 hodgkin's disease, lymphocytic
053 0000097 0.104 neoplasm, benign
053 0000265 0.104 fibromyxosarcoma
053 0001167 0.104 myxoid liposarcoma
053 0000046 0.104 fascial fibrosarcoma
053 0000027 0.104 blue nevus, malignant
053 0000128 0.104 spermatocytic seminoma
053 0007534 0.107 medullary carcinoma nos
053 0000053 0.107 hypereosinophilic syndrome
053 0000042 0.107 odontogenic tumor, malignant
053 0000780 0.107 granulosa cell tumor, malignant
053 0000350 0.107 malignant melanoma in junctiona
054 0000032 0.107 neuroma nos
054 0000025 0.107 balloon cell melanoma
054 0000135 0.107 malignant mastocytosis
054 0000142 0.107 mesonephroma, malignant
054 0003757 0.108 mucoepidermoid carcinoma
054 0010414 0.111 lobular carcinoma in situ
054 0000029 0.111 thymoma, type b2, malignant
054 0000117 0.111 nk/t-cell lymphoma, nasal and n
054 0000890 0.111 anaplastic large cell lymphoma,
055 0000614 0.111 chordoma
055 0000023 0.111 angiomyosarcoma
055 0000111 0.111 fibrous meningioma
055 0000959 0.112 thymoma, malignant
055 0000328 0.112 adenocarcinoid tumor
055 0000057 0.112 chromophobe carcinoma
055 0000032 0.112 megakaryocytic myelosis
055 0000903 0.112 endometrial stromal sarcoma
055 0000120 0.112 atypical medullary carcinoma
055 0000265 0.112 mucocarcinoid tumor, malignant
055 0000070 0.112 juvenile carcinoma of the breas
055 0000029 0.112 adenocarcinoma in situ in famil
056 0000042 0.112 gliomatosis cerebri
056 0006891 0.114 comedocarcinoma nos
056 0000028 0.114 theca cell carcinoma
056 0000096 0.114 myxoid leiomyosarcoma
056 0048315 0.128 malignant melanoma nos
056 0000030 0.128 angiomatous meningioma
056 0000144 0.128 transitional meningioma
056 0000045 0.128 thymoma, type ab, malignant
056 0000140 0.128 pleomorphic rhabdomyosarcoma
056 0000412 0.128 malignant melanoma, regressing
056 0003428 0.129 mucinous cystadenocarcinoma nos
056 0000086 0.129 papillary carcinoma, columnar c
056 0000485 0.129 papillary mucinous cystadenocar
056 0002754 0.130 intraductal and lobular in situ
056 0000626 0.130 hodgkin's disease, lymphocytic
056 0000039 0.130 endometrioid adenocarcinoma, se
056 0000147 0.130 primary cutan. cd30+ t-cell lym
057 0000089 0.130 myxosarcoma
057 0000403 0.130 adenosarcoma
057 0021999 0.137 melanoma in situ
057 0000855 0.137 islet cell carcinoma
057 0000145 0.137 mixed type liposarcoma
057 0003285 0.138 inflammatory carcinoma
057 0000066 0.138 thymoma, type b1, malignant
057 0000047 0.138 spindle cell melanoma, type a
057 0000026 0.138 subcutaneous panniculitis-like
058 0000011 0.138 vipoma
058 0000202 0.138 myeloid sarcoma
058 0002147 0.138 hairy cell leukemia
058 0000191 0.139 stromal sarcoma, nos
058 0000038 0.139 insulinoma, malignant
058 0000032 0.139 glucagonoma, malignant
058 0006684 0.140 adenocarcinoma in situ
058 0001435 0.141 oxyphilic adenocarcinoma
058 0001770 0.141 acute monocytic leukemia
058 0000054 0.141 malignant myoepithelioma
058 0003319 0.142 adenoid cystic carcinoma
058 0000079 0.142 thymoma, type b3, malignant
058 0000477 0.142 spindle cell melanoma, type b
058 0000229 0.142 meningotheliomatous meningioma
058 0000234 0.143 malignant tumor, small cell typ
058 0009679 0.145 comedocarcinoma, noninfiltratin
058 0000207 0.145 cyst-associated renal cell carc
058 0000843 0.146 intraductal micropapillary carc
058 0004480 0.147 ml, large b-cell, diffuse, immu
058 0008089 0.149 squamous cell carcinoma, large
059 0003124 0.150 sarcoma nos
059 0008070 0.152 leiomyosarcoma nos
059 0000114 0.152 atypical meningioma
059 0000082 0.152 thymic carcinoma, nos
059 0000016 0.152 aggressive nk-cell leukemia
059 0002839 0.153 cribriform carcinoma in situ
059 0027965 0.161 papillary adenocarcinoma nos
059 0000018 0.161 malignant eccrine spiradenoma
059 0001301 0.161 papillary cystadenocarcinoma no
059 0000050 0.161 solitary fibrous tumor, maligna
059 0000091 0.161 polymorphous low grade adenocar
059 0004086 0.163 adenocarcinoma with squamous me
059 0000448 0.163 acute myeloid leukemia without
059 0035630 0.173 intraductal carcinoma, noninfil
059 0000022 0.173 acute myeloid leukemia, 11q23 a
059 0000020 0.173 mixed islet cell and exocrine a
059 0004558 0.174 infiltr. duct mixed with other
060 0008688 0.176 nodular melanoma
060 0000030 0.176 insular carcinoma
060 0003377 0.177 mycosis fungoides
060 0000764 0.178 amelanotic melanoma
060 0001025 0.178 spindle cell sarcoma
060 0000113 0.178 queyrat's erythroplasia
060 0000401 0.178 epithelioid cell melanoma
060 0016028 0.183 carcinoid tumor, malignant
060 0000486 0.183 epithelioid leiomyosarcoma
060 0001267 0.183 mature t-cell lymphoma, nos
060 0001045 0.183 cutaneous t-cell lymphoma, nos
060 0000020 0.183 carcinosarcoma, embryonal type
060 0001669 0.184 duct carcinoma in situ, solid t
060 0001021 0.184 liposarcoma, well differentiate
060 0000014 0.184 granulosa cell-theca cell tumor
060 0000614 0.184 acute myeloid leukemia with mat
060 0000617 0.184 renal cell carcinoma, chromopho
060 0000166 0.185 carcinoid tumor, argentaffin, m
060 0000126 0.185 adult t-cell leukemia/lymphoma
060 0000299 0.185 squamous cell carcinoma, small
060 0008938 0.187 malignant lymphoma, follicular
061 0019907 0.193 glioblastoma nos
061 0000906 0.193 meningioma, malignant
061 0000015 0.193 epithelioma, malignant
061 0026705 0.201 endometrioid carcinoma
061 0018131 0.206 acute myeloid leukemia
061 0010280 0.209 chronic myeloid leukemia
061 0000026 0.209 polygonal cell carcinoma
061 0000057 0.209 collecting duct carcinoma
061 0000303 0.209 metaplastic carcinoma, nos
061 0000382 0.209 mixed tumor, malignant nos
061 0333623 0.303 infiltrating duct carcinoma
061 0002006 0.303 acute myelomonocytic leukemia
061 0020557 0.309 clear cell adenocarcinoma nos
061 0000885 0.309 infiltrating ductular carcinoma
061 0000165 0.309 carcinoma in pleomorphic adenom
061 0000661 0.310 mixed epithel. & spindle cell m
061 0000295 0.310 glioblastoma with sarcomatous c
061 0000042 0.310 adenocarcinoma with apocrine me
061 0021695 0.316 infiltrating duct and lobular c
061 0000324 0.316 acute myeloid leukemia, minimal
061 0000100 0.316 papillary squamous cell carcino
061 0000012 0.316 endometrioid adenocarcinoma, ci
061 0000074 0.316 hodgkin's disease, lymphocytic
061 0002217 0.317 paget's disease and infiltratin
062 0001323 0.317 liposarcoma nos
062 0006337 0.319 tubular adenocarcinoma
062 0000014 0.319 heavy chain disease, nos
062 0000077 0.319 atypical carcinoid tumor
062 0000494 0.319 skin appendage carcinoma
062 0001092 0.319 mixed cell adenocarcinoma
062 0001281 0.320 spindle cell melanoma nos
062 0004682 0.321 serous cystadenocarcinoma nos
062 0005812 0.323 fibrous histiocytoma, malignant
062 0001263 0.323 gastrointestinal stromal sarcom
062 0000106 0.323 malignant tumor, giant cell typ
062 0000493 0.323 basaloid squamous cell carcinom
062 0000258 0.323 renal cell carcinoma, sarcomato
062 0000118 0.323 epithelial-myoepithelial carcin
062 0000884 0.323 acral lentiginous melanoma, mal
062 0015063 0.328 papillary serous cystadenocarci
062 0000039 0.328 endometrioid adenofibroma, mali
062 0012432 0.331 malignant lymphoma, non hodgkin
062 0000037 0.331 adenocarcinoma with spindle cel
062 0000086 0.331 therapy-related myelodysplastic
062 0002593 0.332 infiltr. duct mixed with other
062 0003162 0.333 noninfiltrating intraductal pap
062 0005812 0.334 malignant lymphoma, mixed lymph
062 0003414 0.335 malignant lymphoma, follicular
063 0001353 0.336 hemangiosarcoma
063 0000023 0.336 hodgkin's sarcoma
063 0000024 0.336 meningiomatosis nos
063 0000239 0.336 solid carcinoma nos
063 0000135 0.336 composite carcinoid
063 0001142 0.336 cystadenocarcinoma nos
063 0000023 0.336 schneiderian carcinoma
063 0011193 0.339 adenosquamous carcinoma
063 0000084 0.339 psammomatous meningioma
063 0037088 0.350 ml, large b-cell, diffuse
063 0000080 0.350 basal cell adenocarcinoma
063 0000016 0.350 intestinal t-cell lymphoma
063 0000287 0.350 dedifferentiated liposarcoma
063 0000939 0.350 plasmacytoma, extramedullary
063 0000869 0.350 plasma cell tumor, malignant
063 0003345 0.351 malignant lymphoma, nodular nos
063 0001358 0.352 paget disease and intraductal c
063 0002708 0.353 serous surface papillary carcin
063 0000020 0.353 squamous cell carcinoma, clear
063 0000012 0.353 basal cell carcinoma, fibroepit
063 0000334 0.353 giant cell sarcoma (except of b
063 0023109 0.359 squamous cell carcinoma, kerati
063 0000472 0.359 infiltrating lobular mixed with
063 0000277 0.359 combined hepatocellular carcino
064 0001275 0.360 polycythemia vera
064 0000041 0.360 lymphangiosarcoma
064 0017866 0.365 oat cell carcinoma
064 0037543 0.375 renal cell carcinoma
064 0001065 0.376 giant cell carcinoma
064 0011520 0.379 acinar cell carcinoma
064 0001358 0.379 cloacogenic carcinoma
064 0034336 0.389 lobular carcinoma nos
064 0013199 0.393 malignant lymphoma nos
064 0000947 0.393 granular cell carcinoma
064 0000405 0.393 pleomorphic liposarcoma
064 0000723 0.393 apocrine adenocarcinoma
064 0003164 0.394 scirrhous adenocarcinoma
064 0005106 0.396 neuroendocrine carcinoma
064 0000363 0.396 sweat gland adenocarcinoma
064 0247826 0.465 squamous cell carcinoma nos
064 0019957 0.471 hepatocellular carcinoma nos
064 0000757 0.471 papillary squamous cell carcino
064 0010254 0.474 carcinoma, undifferentiated typ
064 0000087 0.474 acute panmyelosis with myelofib
064 0000248 0.474 giant cell and spindle cell car
064 0000083 0.474 small cell carcinoma, fusiform
064 0000151 0.474 adenocarcinoma in mult. adenoma
064 0000018 0.474 atypical chronic myeloid leuk.,
064 0000026 0.474 adenocarcinoma with cartilagino
065 0002723 0.475 meningioma nos
065 0003731 0.476 acute leukemia nos
065 0000880 0.476 cribriform carcinoma
065 0000200 0.477 plasma cell leukemia
065 0000514 0.477 pleomorphic carcinoma
065 0000052 0.477 eccrine adenocarcinoma
065 0029592 0.485 large cell carcinoma nos
065 0000123 0.485 brenner tumor, malignant
065 0000998 0.485 essential thrombocythemia
065 0000016 0.485 ceruminous adenocarcinoma
065 0010967 0.488 signet ring cell carcinoma
065 0000125 0.488 nodular hidradenoma, malignant
065 0004446 0.490 carcinoma, anaplastic type nos
065 0000086 0.490 adenoid squamous cell carcinoma
065 0000116 0.490 sclerosing sweat duct carcinoma
065 0000856 0.490 adenocarcinoma with mixed subty
065 0003638 0.491 marginal zone b-cell lymphoma,
065 0004319 0.492 ml, mixed sm. and lg. cell, dif
065 0000183 0.492 superficial spreading adenocarc
065 0000054 0.492 hepatocellular carcinoma, clear
065 0000020 0.492 composite hodgkin and non-hodgk
065 0000105 0.492 adenocarcinoma with neuroendocr
065 0001094 0.493 intraductal papillary adenocarc
066 0000815 0.493 erythroleukemia
066 0001654 0.493 linitis plastica
066 0000461 0.493 basaloid carcinoma
066 0002890 0.494 mantle cell lymphoma
066 0001270 0.495 ml, lymphoplasmacytic
066 0000787 0.495 spindle cell carcinoma
066 0001267 0.495 carcinoma, diffuse type
066 0000390 0.495 alveolar adenocarcinoma
066 0000618 0.496 paget's disease, mammary
066 0050173 0.510 small cell carcinoma nos
066 0000624 0.510 papillary carcinoma in situ
066 0000783 0.510 desmoplastic melanoma, malignan
066 0011881 0.513 bronchiolo-alveolar adenocarcin
066 0000241 0.513 angioimmunoblastic t-cell lymph
066 0000403 0.514 large cell neuroendocrine carci
066 0000315 0.514 malignant tumor, fusiform cell
066 0000042 0.514 prolymphocytic leukemia, t-cell
066 0001669 0.514 small cell carcinoma, intermedi
066 0000050 0.514 adenocarc. in situ in mult. ade
066 0000127 0.514 neoplasm, uncertain whether ben
066 0000032 0.514 intraductal papillary-mucinous
067 0000114 0.514 sezary's disease
067 0001015 0.515 lymphoid leukemia nos
067 0000779 0.515 mesodermal mixed tumor
067 0000184 0.515 trabecular adenocarcinoma
067 0000514 0.515 intracystic carcinoma, nos
067 0009635 0.518 ml, small b lymphocytic, nos
067 0000557 0.518 combined small cell carcinoma
067 0025451 0.525 mucin-producing adenocarcinoma
067 0000032 0.525 dedifferentiated chondrosarcoma
067 0000016 0.525 immunoproliferative disease, no
067 0001263 0.525 epithelioid mesothelioma, malig
067 0000179 0.525 splenic marginal zone b-cell ly
067 0000212 0.525 bronchiolo-alveolar carcinoma,
067 0000584 0.526 squamous cell carcinoma, spindl
067 0008163 0.528 adenocarcinoma in situ in adeno
067 0000165 0.528 bronchiolo-alveolar carcinoma,
067 0005831 0.530 adenocarcinoma in situ in tubul
067 0000327 0.530 acute myeloid leuk. with multil
067 0000026 0.530 bronch.-alv. carc., mixed mucin
067 0000038 0.530 intraductal papillary-mucinous
068 0000054 0.530 carcinoma simplex
068 0002264 0.530 carcinosarcoma nos
068 1021940 0.818 adenocarcinoma nos
068 0003088 0.819 mullerian mixed tumor
068 0000758 0.819 villous adenocarcinoma
068 0045778 0.832 mucinous adenocarcinoma
068 0004897 0.834 mesothelioma, malignant
068 0001673 0.834 verrucous carcinoma nos
068 0014878 0.838 non-small cell carcinoma
068 0000032 0.838 thymoma, type a, malignant
068 0000295 0.838 pseudosarcomatous carcinoma
068 0000056 0.838 granular cell tumor, malignant
068 0018652 0.844 hutchinson's melanotic freckle
068 0020880 0.849 adenocarcinoma in adenomatous p
068 0000032 0.849 prolymphocytic leukemia, b-cell
068 0096537 0.877 papillary transitional cell car
068 0016555 0.881 adenocarcinoma in tubulovillous
069 0036429 0.892 multiple myeloma
069 0004610 0.893 cholangiocarcinoma
069 0001872 0.893 myeloid leukemia nos
069 0000156 0.893 basosquamous carcinoma
069 0000046 0.893 eccrine poroma, malignant
069 0000010 0.893 clear cell adenocarcinofibroma
069 0000476 0.894 fibrous mesothelioma, malignant
069 0016754 0.898 adenocarcinoma in villous adeno
069 0000940 0.899 transitional cell carcinoma in
069 0000419 0.899 noninfiltrating intracystic car
069 0000362 0.899 myelosclerosis with myeloid met
069 0000586 0.899 chronic myeloproliferative dise
069 0004672 0.900 adenocarcinoma in situ in villo
069 0002438 0.901 papillary trans. cell carcinoma
069 0007272 0.903 malignant melanoma in hutchinso
070 0003575 0.904 tumor cells, malignant
070 0030328 0.913 chronic lymphoid leukemia
070 0000301 0.913 prolymphocytic leukemia, nos
070 0050243 0.927 transitional cell carcinoma nos
070 0000049 0.927 osteosarcoma in paget's disease
071 0001585 0.927 waldenstrom macroglobulinemia
071 0000122 0.927 transitional cell carcinoma, sp
071 0000263 0.927 refractory cytopenia with multi
071 0000080 0.927 refract. anemia with excess bla
071 0000811 0.928 paget's disease, extramammary (
072 0002581 0.928 leukemia nos
072 0182616 0.980 carcinoma nos
072 0000408 0.980 klatskin tumor
072 0000012 0.980 hepatoid adenocarcinoma
072 0000796 0.980 sebaceous adenocarcinoma
072 0000786 0.980 basal cell carcinoma nos
072 0000012 0.980 adenoid basal cell carcinoma
072 0002766 0.981 adenocarcinoma, intestinal type
072 0000015 0.981 multicentric basal cell carcino
072 0000763 0.981 refractory anemia with excess b
072 0000261 0.981 mesothelioma, biphasic type, ma
072 0000028 0.981 transitional cell carcinoma, mi
073 0000820 0.982 refractory anemia
073 0000036 0.982 basal cell carcinoma, nodular
074 0001716 0.982 merkel cell carcinoma
074 0002422 0.983 myelodysplastic syndrome, nos
074 0000655 0.983 refractory anemia with siderobl
074 0001798 0.984 chronic myelomonocytic leukemia
074 0000089 0.984 myelodysplastic syndr. with 5q
076 0056558 1.000 neoplasm, malignant
As specified in the Limited-Use Data Agreement, the citation for the SEER data
is as follows:
Surveillance, Epidemiology, and End Results (SEER) Program
(www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer
Institute, DCCPS, Surveillance Research Program, Cancer Statistics
Branch, released April 2008, based on the November 2007 submission.
Once we have the columned data, we can easily produce a graphic that represents the salient features we would like to emphasize.

In the next few blogs, I'll explain the clinical significance of this little demo project, and I'll describe the free, open source, techniques that I used to extract and compile the data. Afterwards, I'll show you how we can drill into the data to refine the questions we can ask, so that we can draw conclusions that can stand up to critical inspection.
For Perl and Ruby programmers, methods and scripts for using SEER and other publicly available biomedical databases, are described in detail in my prior books:
Perl Programming for Medicine and Biology
Ruby Programming for Medicine and Biology
- © 2008 Jules Berman
As specified in the SEER Data Agreement, the citation for the SEER data is as follows:
"Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Limited-Use Data (1973-2005), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2008, based on the November 2007 submission."
As with all of my scripts, lists, web sites, and blog entries, the following disclaimer applies. This material is provided by its creator, Jules J. Berman, "as is", without warranty of any kind, expressed or implied, including but not limited to the warranties of merchantability, fitness for a particular purpose and noninfringement. in no event shall the author or copyright holder be liable for any claim, damages or other liability, whether in an action of contract, tort or otherwise, arising from, out of or in connection with the material or the use or other dealings in the material.
In June, 2014, my book, entitled Rare Diseases and Orphan Drugs: Keys to Understanding and Treating the Common Diseases was published by Elsevier. The book builds the argument that our best chance of curing the common diseases will come from studying and curing the rare diseases.
I urge you to read more about my book. There's a generous preview of the book at the Google Books site. If you like the book, please request your librarian to purchase a copy of this book for your library or reading room.
Subscribe to:
Posts (Atom)
