Today, I want to go back and review the relationship between RDF Schemas and directed graphs and to provide a rationale for why this is possible and to provide you with some tools you might need to do this for your own RDF Schemas.
RDF (Resource Description Framework) is a syntax and logic for making computer parsable statements that have meaning. RDF uses the familiar tagging syntax of XML, so all RDF documents are also XML documents.
Meaning, in the informatics field, is achieved when a datum and a description of the datum (a metadatum) are bound to an identified object. This is the "triple" that underlies all of RDF.
Here is a data/metadata pair:
<date>March 15, 2008</data>
This kind of data/matadata pair, common to XML, has no meaning because it is not bound to an identified object.
Now consider:
Ides of March occurs on <date>March 15, 2008</data>
This gets a little closer to an RDF statement (with meaning) because the data/metadata pair are assigned to an object (Ides of March).
RDF has a formal way of defining objects [and their properties, which we won't discuss here]. This is called RDF Schema. You can think of RDF Schema as a dictionary for the terms in an RDF data document. RDF Schema is written in RDF syntax. This means that all RDF Schemas are RDF documents and consist of statments in the form of triples.
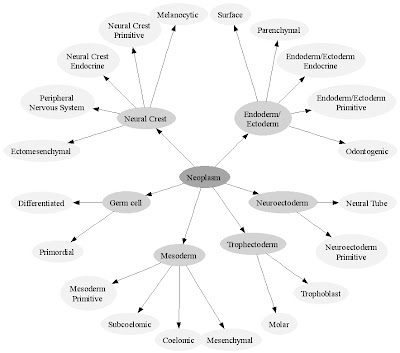
For today, the important point about RDF Schemas is that they create logical relationships among objects in a domain that can be translated into directed graphs (graphs consisting of connected nodes and arcs and directions for the arcs).
An example of two RDF statements:
<rdfs:Class rdf:ID="Fibrous_tissue">
<rdfs:subClassOf
neo:resource="#Connective_tissue"/>
</rdfs:Class>
<rdfs:Class rdf:ID="Mesoderm_primitive">
<rdfs:subClassOf
neo:resource="#Mesoderm"/>
</rdfs:Class>
Every class of object is a subclass of another class of object. A Perl script, such as the one that I provided yesterday, can parse and RDF Schema and transform it into a GraphViz script. This is a type of poor-man's metaprogramming (using a programming language to generate programs in another programming language). GraphViz is a free, open source graphic scripting language that renders a wide range of graphic representations for specified object relationships.
Information on GraphViz is available at:
http://www.graphviz.org/
RDF is the syntax and logic underlying the semantic web, and every serious informatician must learn to use RDF. There are quite a few books and articles written on RDF. My book, Ruby Programming for Medicine and Biology , has a large section on RDF with some examples showing how to build and use RDF Schemas and RDF documents. In my opinion, Ruby is a better language that Perl or Python for dealing with RDF logic. Also, based on my limited ability to survey all of the literature, it would seem that a really good book that explains RDF and provides good examples for building RDF documents and drawing useful inferences from multiple RDF documents, has not been written. When I find one, I'll let you know.
- Jules Berman
key words: RDF Schema, triple, triples, ontology, digraph
Science is not a collection of facts. Science is what facts teach us; what we can learn about our universe, and ourselves, by deductive thinking. From observations of the night sky, made without the aid of telescopes, we can deduce that the universe is expanding, that the universe is not infinitely old, and why black holes exist. Without resorting to experimentation or mathematical analysis, we can deduce that gravity is a curvature in space-time, that the particles that compose light have no mass, that there is a theoretical limit to the number of different elements in the universe, and that the earth is billions of years old. Likewise, simple observations on animals tell us much about the migration of continents, the evolutionary relationships among classes of animals, why the nuclei of cells contain our genetic material, why certain animals are long-lived, why the gestation period of humans is 9 months, and why some diseases are rare and other diseases are common. In “Armchair Science”, the reader is confronted with 129 scientific mysteries, in cosmology, particle physics, chemistry, biology, and medicine. Beginning with simple observations, step-by-step analyses guide the reader toward solutions that are sometimes startling, and always entertaining. “Armchair Science” is written for general readers who are curious about science, and who want to sharpen their deductive skills.